
Segmentation and fitting wih Segger - Gallery
| GroEL | ||
| Density map | Segmented regions | Fitted structures |
 EMD: 1042 resolution: 10Å threshold: 0.17 |
 14 regions after smoothing with 10 steps of size 5.0Å and sharpening |
 PDB:1xck chain A fitted by alignment to 1 region (the region is shown using a transparent surface)  PDB:1xck chain A fitted by alignment to each of the 14 regions (the structure is shown in each of the 14 resulting fits with a different colour) |
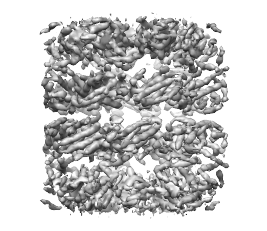 EMD: 5001 resolution: 4.2Å threshold: 0.89 |
 14 regions after smoothing with 4 steps of size 7.5Å and sharpening |
 PDB:1xck chain A fitted by alignment to 1 region (the region is shown using a transparent surface) 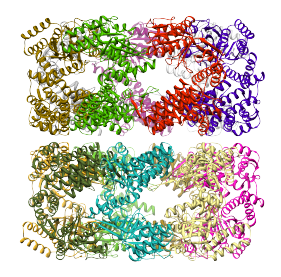 PDB:1xck chain A fitted by alignment to each of the 14 regions (the structure is shown in each of the 14 resulting fits with a different colour) |
| GroEL+GroES | ||
| Density map | Segmented regions | Fitted structures |
 EMD: 1180 resolution: 7.7Å threshold: 0.17 |
 37 regions after smoothing with 2 steps of size 5.0Å and sharpening  21 regions after joining regions corresponding to the same protein |
 PDB:1aon chain O fitted by alignment to 1 region (the region is shown with a transparent surface)  PDB:1aon chain A fitted by alignment to 2 regions (the regions were joined into one region, which is shown using a transparent surface)  PDB:1aon chain H fitted by alignment to 2 regions (the regions were joined into one region, which is shown using a transparent surface) |
 PDB:1aon chains O (blue), A (red), H (green), each fitted 7 times, by alignment to regions as shown above |
||
| Ribosome | ||
| Density map | Segmented regions | Fitted structures |
 EMD: 1056 resolution: 9.0Å threshold: 25 |
 2 regions after smoothing with 9 steps of size 10.0Å and sharpening (green region is the large subunit, and the orange region is the small subunit) |
 PDB:2avy (the entire small subunit, which is also made up of RNA and proteins) the structure was fitted by aligning it with the orange region (the region is shown with a transparent surface)  PDB:2aw4 (the entire large subunit, which is also made up of RNA and proteins) the entire structure was fitted by aligning it with the green region |
 44 regions 43 proteins were fit to regions obtained by segmenting the non-smoothed map the regions that overlapped a fitted protein were joined to create a single region for each protein the remaining regions were joined to create the dark grey region, which is RNA+other proteins |
 PDB:2avy chain B  PDB:2avy chain J  PDB:2avy chain M  PDB:2aw4 chain G  PDB:2aw4 chain P The proteins above are only 5 of the 49 proteins in the large and small ribosome subunits combined. These were correctly fitted by aligning the structures with groups of regions. 38 of the other proteins were also fitted correctly, but are not shown here. |
|
| Bacteriophage Lambda Capsid | ||
| Density map | Segmented regions | Fitted structures |
 EMD: 1507 resolution: 14.5Å threshold: 3.5 |
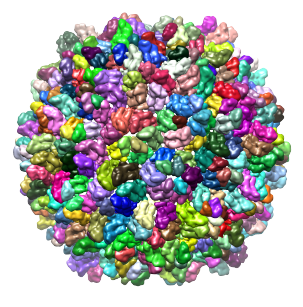 353 regions after smoothing with 1 step of size 10.0Å and sharpening The capsid is a T=7 icosahedral lattice, built from 60 asymmetric units that contain 7 proteins each. |
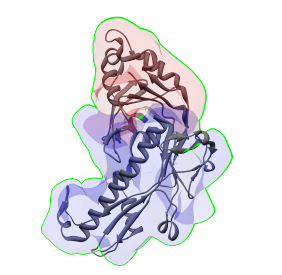 PDB:3bqw The structure of a single protein was aligned to two of the regions, which are shown above using transparrent surfaces. |
 10 regions that make up an asymmetric unit (ASU) 4 of the regions correspond to single proteins, and the other 6, taken in groups of two, correspond to 1 protein per group. |
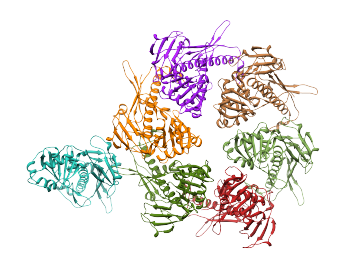 The structure of a single protein was aligned with single or groups of two regions, recreating the structure of a single ASU. |
|
Please contact Greg Pintilie by email with comments and suggestions.

