
download all solution code here:
http://people.csail.mit.edu/~emax/6.869/ps1/max-ps1.code.zip
function [alpha, beta, theta, u0, v0, R, t] = calibrate(f2D,f3D) %% %% computes the intrinsic (camera) and extrinsic (world) parameters %% given real-world coordinates in homogenous coords (f3D) of some %% item in space, and corresponding pixel locations in f2d %% M = projM(f2D,f3D) % first estimate the projection matrix % M = (A b) A = M(:,[1:3]); b = M(:,4); % then recover the parameters a1 = A(1,:)'; a2 = A(2,:)'; a3 = A(3,:)'; % eqtns 3.11 in forsythe + ponce rho = 1/sqrt(sum(a3.^2)) r3 = rho * a3 u0 = rho.^2 * (a1' * a3) v0 = rho.^2 * (a2' * a3); % equations 3.13 in forsythe + ponce theta = acos( -(cross(a1, a3)'*cross(a2,a3)) / ( sqrt(sum(cross(a1,a3).^2)) * sqrt(sum(cross(a2,a3).^2)) ) ); alpha = rho.^2 * sqrt(sum(cross(a1,a3).^2))*sin(theta); beta = rho.^2 * sqrt(sum(cross(a2,a3).^2))*sin(theta); % eqtns 3.14 in forsythe + ponce r1 = rho ^2 * sin(theta) / beta * cross(a2,a3); r2 = cross(r3,r1); R = [r1'; r2'; r3'] K = [alpha -alpha*cot(theta) u0; 0 beta/sin(theta) v0; 0 0 1]; %rho * A * inv(R); t = rho * inv(K)*b;results:
[alpha beta theta u0 v0 R t] = calibrate(f2D,f3D);
alpha =
1.1177e+03
beta =
1.1177e+03
theta =
1.5708
u0 =
300.5000
v0 =
300.4991
R =
0.7071 0.0000 -0.7071
-0.4150 0.8097 -0.4150
0.5725 0.5869 0.5725
t =
-0.0000
0.0810
-6.9277
function f2D = pcamview(f3D, alpha, beta, theta, u0, v0, R, t) % projects f3D onto a plane according to the given extrinsic % and intrinsic parameters f2D = []; for p = f3D, %% from 2.17 in forsythe and ponce M(1,[1:3]) = alpha*R(1,:) - alpha * cot(theta) * R(2,:) + u0 * R(3,:); M(2,[1:3]) = beta/sin(theta)*R(2,:) + v0*R(3,:); M(3,[1:3]) = R(3,:); M(1,4) = alpha*t(1) - alpha*cot(theta)*t(2) + u0*t(3); M(2,4) = beta/sin(theta)*t(2) + v0*t(3); M(3,4) = t(3); %% from 2.16 in forsythe and ponce z = M(3,:)*p u = (M(1,:)*p)/(z) v = (M(2,:)*p)/(z) f2D = [f2D [u v]']; endresult:
>> pcamview(f3D,alpha,beta,theta,u0,v0,R,t) ans = 300.5000 574.0207 300.5000 490.9958 300.5000 396.3433 300.5000 287.4363 222.4527 512.7543 217.6516 430.9434 212.2211 338.4075 206.0288 232.8895 152.7584 458.0450 144.1846 377.6910 134.5544 287.4362 123.6596 185.3304 90.1441 408.8933 78.5908 330.1454 65.6946 242.2448 51.2069 143.4970 549.7931 143.4970 477.3404 185.3304 394.9712 232.8895 457.2591 106.4140 383.5976 143.4970 300.5000 185.3304 374.6685 73.3157 300.5000 106.4140 217.4024 143.4970 300.5000 43.5927 226.3315 73.3157 143.7409 106.4140 535.3054 242.2448 466.4456 287.4362 388.7789 338.4075 522.4092 330.1454 456.8154 377.6910 383.3484 430.9434 510.8559 408.8933 448.2416 458.0450 378.5473 512.7543

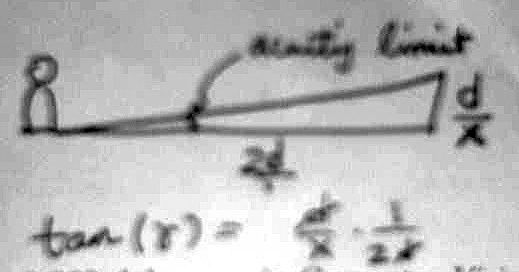
The maximum acuity according to the acutiy limit equation (a=0.23e+0.7) would be when the eccentricity, e=0. At this point, we get an acuity of 0.7. Half of the acuity here would correspond to twice this value, or 1.4. Using simple trigonometry, we have an image with X pixels per side, adding up to a length D, and stand from a distance 2D. The angle that the image makes with our eye occurs at half the acuity limit, 1.4 minutes, or 1.4*1/60 degrees. Therefore we have the relation
tan (1.4*1/60) = d/x * 1/(2d)the d's cancel, leaving us:
x = 1/(2*tan (1.4*1/60)) x = 2455/2 = 1228 pixels per side
e(i,j) = atan(sqrt( ((i/X).^2 +(j/X)^2))/2 )where X represents the number of pixels per side. Now, we need to compute d_view, which is the distance to the point at which we are looking
d_view = sqrt(4 + (i/X)^2 + (j/X).^2 )Now, d_spacing is simply the arc inscribed by the angle corresponding to the limit of half the maximum visual acuity, given at a distance of d_view.
d_spacing = d_view * tan((0.23*(e(i/j)*180/PI)+0.7) * 1/60 * pi/180))The scale factors (1/60*pi/180) above need to exist in order to convert the "minutes" values back to radians.
2^n/X = d_view * tan((0.23*(e(i/j)*180/PI)+0.7) * 1/60 * pi/180))yields:
n = log2(X * d_view * tan((0.23*(e(i/j)*180/PI)+0.7) * 1/60 * pi/180))I implemented this as a function in matlab as follows:
function n = nlevels(x, y, WIDTH) % computes number of gaussian pyramid levels we need for problem 2 d sx = x/WIDTH; sy = y/WIDTH; e = atan(0.5*sqrt(sx^2+sy^2)); a = 0.23 * e*(180/pi) + 1.4; dview = sqrt(sx^2 + sy^2 + 4); n = log2(WIDTH * dview * tan (a/60 * pi/180));
result:

function synthIM = SynthTexture(sample, w, s)
% original texture sample
% initialize to a negative number so it's easy to distinguish between
% filled and unfilled
synthIM = -1*ones(s);
synthSize = s;
%% ====================== utility funcitons ========================
function b = isfilled(ijs)
idxs = sub2ind(synthSize,ijs(:,1),ijs(:,2));
b = ~(synthIM(idxs)==-1);
end
%% == findEmpty ==
function plist = findEmpty(synthIM, w)
% finds boundary entries that need to be filled
%
% try converting to binary first then xor the difference
simsize = size(synthIM);
dilate = imdilate(synthIM,ones(3));
dilthresh = threshzero(dilate);
synthresh = threshzero(synthIM);
edges = xor ( dilthresh, synthresh );
ptlst = find(edges == 1.0);
[is js] = ind2sub(simsize,ptlst);
is = is - floor(w/2); js = js - floor(w/2);
plist = [is'; js'];
%% delete out of bounds points
%% plist(:,min(plist,[],1)<=0)=[];
end
== end of findEmpty ==
== ValidMatrix.m ==
function validMatrix = makeValidMatrix(Sample,i,j,width)
%%
%% Constructs a ValidMatrix for a sample with upper-left
%% corner (i,j) and width width
validMatrix = ones(width);
sample = getSample(Sample,i,j,width);
for ii=i:i+width-1,
for jj=j:j+width-1,
vi = ii-i+1;
vj = jj-j+1;
%% boundary conditions
if ( ii < 1 | jj < 1 | ii > size(Sample,1) | jj > size(Sample,2) )
validMatrix(vi,vj)=0;
end
if(sample(vi,vj)<0)
validMatrix(vi,vj)=0;
end
end
end
%% == end of makeValidMatrix ==
%% == SynthTexture main ==
%% pick a random wxw sample from sample and plop it in the middle
seedw = 3;
si = s(1)/2; sj = s(2)/2;
rsrci = 32 - floor(seedw/2); %-floor(w/2); % floor(rand(1)*(size(sample,1)-seedw))+1;
rsrcj = 4 - floor(seedw/2); %-floor(w/2); % floor(rand(1)*(size(sample,2)-seedw))+1;
synthIM(si:si+seedw-1,sj:sj+seedw-1) = ...
sample(rsrci:rsrci+seedw-1,rsrcj:rsrcj+seedw-1);
figure(4);
testshow(synthIM);
figure(1);
%% =================================================================
% precompute a gaussian mask
sigma = w/6.4;
gmask = gauss2d(w,sigma);
% find things to fill
tofill = findEmpty(synthIM,w)
counter = 0;
%% debugging
disposed = 0;
%% end debugging
wmiddle = floor(w/2);
while ( length(find(synthIM<0)) > 0) % while we have unfilled pixels
lengthtofill = length(tofill)
for pt = tofill,
%% what to do %%
pt_i = pt(1); pt_j = pt(2);
template = getSample(synthIM,pt_i,pt_j,w);
validmask = makeValidMatrix(synthIM,pt_i,pt_j,w);
G = validmask .* gmask;
[bestmatches err] = findMatches(template,sample,G);
% select one
if (length(bestmatches) > 0)
pick = floor(size(bestmatches,3)*rand(1))+1; %% pick one at random!
bestmatch = bestmatches(:,:,pick);
synthIM(pt_i + wmiddle, pt_j+wmiddle) = bestmatch(wmiddle+1,wmiddle+1);
counter = counter+1;
end
end
tofill = findEmpty(synthIM,w);
end
end
findMatches:
function [bestmatches, errors] = findMatches(template,sample,G)
% searches sample for template and returns the
epsilon = 0.1;
sizeG = size(G);
sizetemplate = size(template);
w = size(template,1);
v = size(template,2);
%sigma = w/6.4;
%gmask = gauss2d(w,sigma);
total = sum(sum(G)); % find maximum by summing over the mask
SSD = [];
for i=1:(size(sample,1) - w),
for j=1:(size(sample,2) - v),
SSD(i,j) = sum(sum( ...
((template-sample([i:i+w-1],[j:j+v-1])).^2 ) .* G));
end
SSD(i,j) = SSD(i,j)/total;
end
idxs = [];
while (length(idxs) == 0)
% minSSD cutoff --
minSSD = min(SSD(:));
idxs = find(SSD < min( DELTA, minSSD*(1+EPSILON)));
EPSILON = EPSILON*1.1;
DELTA=DELTA*1.1;
end
bestmatches = [];
errors = [];
if (length(idxs) > 0)
[i j] = ind2sub(size(SSD),idxs);
for match=1:length(i)
i_match = i(match); j_match = j(match);
bestmatches(:,:,match) = ...
sample([i_match:i_match+w-1],[j_match:j_match+v-1]);
errors(match) = SSD(i_match,j_match);
end
% bestmatches = sample([i:i+w-1],[j:j+v-1]);
end
results:
Each time we run the algorithm, even with the same window size, we will
get different patterns due ot the random process we are employing
to pick the best texture. Here are the set of pictures generated with
EPSILON=0.001, which enforces most strict (optimal) choosing.
starting with seed at (4,32), generating [100 100]:
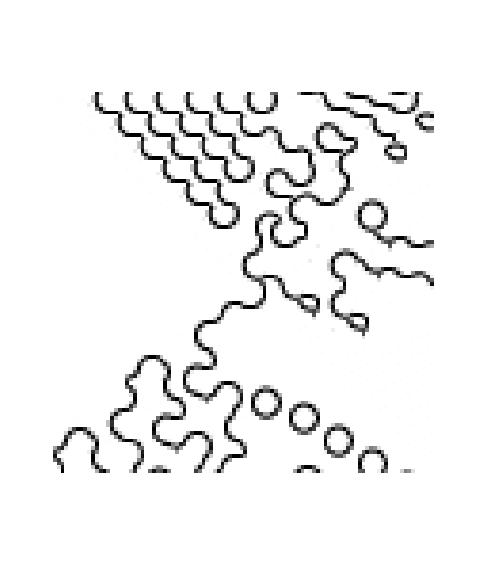
|
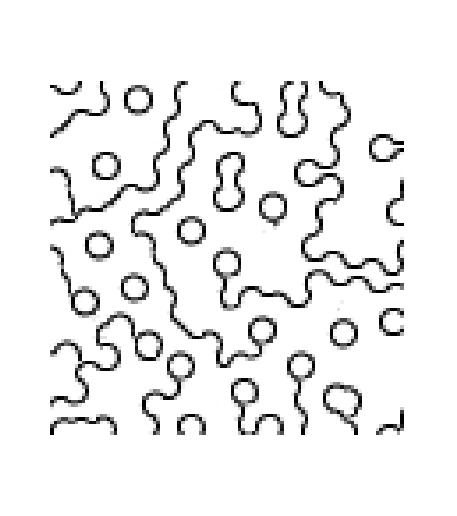
|
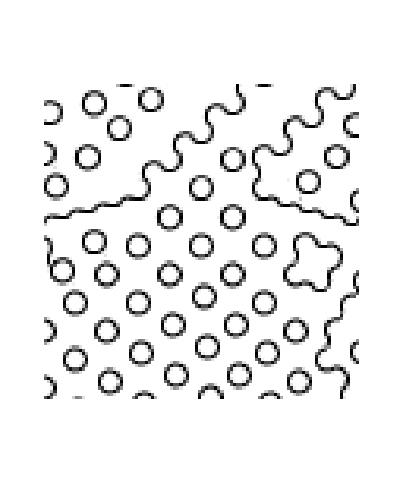
|
>> XYZ = [cx'; cy'; cz']*sA
ans =
9.8584
11.6454
11.7413
X -> 9.85
Y -> 11.64
Z -> 11.74
normalized coefficients X = x / (x + y + z)
Xnorm = XYZ(1)/sum(XYZ) = 0.2965
Ynorm = XYZ(2)/sum(XYZ) = 0.3503
Znorm = XYZ(3)/sum(XYZ) = 0.3532
Given a signal represented with primaries P(450), P(550), P(650)
represent them in P(460) P(510) P(590)
C = [cx'; cy'; cz'];
% establish the primaries
P = zeros(length(cx),3);
P((450-400)/5,1)=1;
P((550-400)/5,2)=1;
P((650-400)/5,3)=1;
e = C*P'*[a b c]';
% now, to find what combination we need in our new basis, compute C*Pnew
% where Pnew is our new basis
Pnew = zeros(length(cx),3);
Pnew((460-400)/5,1)=1;
Pnew((510-400)/5,2)=1;
Pnew((590-400)/5,3)=1;
C*Pnew*enew = C*P*e';
enew = inv(C*Pnew)*C*P*[a b c]';
this matrix, inv(C*Pnew)*C*P has value:
>> inv(C*Pnew)*C*P
1.0327 -0.1849 0.0456
-0.0878 1.5790 -0.3771
0.0196 0.4239 0.3548
So what we are trying to accomplish is to express each of the X, Y, Z primaries as linear combinations of our own Primaries, P(460), P(510), and P(640). Therefore, what we'd like to do is this: eX = w1*eP1 + w2*eP2 + w3*eP3 eY = w4*eP1 + w5*eP2 + w6*eP3 eZ = w7*eP1 * w8*eP2 + w9*eP3 or, more compactly [eX eY eZ]' = W * [eP1 eP2 eP3]' We want to find W. Switching things around, we can write: [eP1 eP2 eP3]' = inv(W) * [eX eY eZ]'; Fortunately, we already know inv(W), since this is the problem of representing an arbitary color (in this case our primaries) as combinations of X, Y, and Z. inv(W) = [cx'; cy'; cz']*[P1 P2 P3]; where P1 = [ 0 0 ... 1 .. 0 ]'; corresponding to wavelength 450
where P2 = [ 0 0 ... 0 .. 1 .. 0 ]'; corresponding to wavelength 510
where P3 = [ 0 0 ... 0 .. 0 .. 1 .. 0 ]'; corresponding to wavelength 640
thus, inv(W) = [Cx(460) Cy(460) Cz(460); Cx(510) Cy(510) Cz(510); Cx(640) Cy(640) Cz(640)]; inverting this back, we getinv(inv(W)) = Wtherefore, we have the relation [eX eY eZ]' = inv([cx';cy';cz']*[P1 P2 P3]) * [eP1 eP2 eP3]'; the value inv([cx';cy';cz']*[P1 P2 P3]) for CIE XYZ and primaries 450, 510, and 640 are: >> P1 = zeros(length(cx),1); >> P2 = zeros(length(cx),1); >> P3 = zeros(length(cx),1); >> P1((450-400)/5,1) = 1; >> P2((510-400)/5,1) = 1; >> P3((640-400)/5,1) = 1; >> inv([cx'; cy'; cz']*[P1 P2 P3]) ans = 0.1137 -0.2840 0.5435 -0.9548 2.3844 0.1466 1.7765 0.1719 -0.3498