|
|
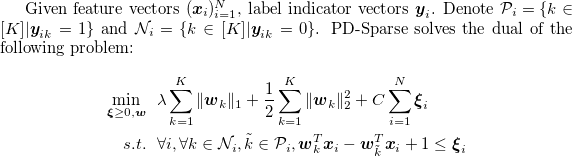
$ git clone https://github.com/a061105/ExtremeMulticlass.git
Three binary files will be generated: multiTrain , multiTrainHash and multiPred. Both multiTrain and multiTrainHash can be used for training since they solve the exact same problem. Note that multiTrainHash is designed to be more memory-efficient whereas multiTrain is faster when memory is sufficient (i.e. when matrices of size (#classes by #features) can fit into memory).
We provide the (train, heldout and test) data sets used in our paper. The data sets are listed below: multilabel data sets: "Eur-Lex", "rcv1_regions", "bibtex", "LSHTCwiki" multiclass data sets: "sector", "aloi.bin", "Dmoz", "LSHTC1", "imageNet" Note the data sets can be also found on other public sites (e.g. libsvm, mulan dataset pages and LSHTC competition sites), but the versions here have been preprocessed (e.g. tf-idf, random features, and scaling). One can download them by $ cd examples/ $ make construct dataset=rcv1_regions Note that the exact names of data sets listed above should be assigned to variable "dataset".
One can train by either of the two commands:
$ ./multiTrain (options) [train_data] (model)
$ ./multiTrainHash (options) [train_data] (model)
options::
-s solver: (default 1)
0 -- Stochastic Block Coordinate Descent
1 -- Stochastic-Active Block Coordinate Descent (PD-Sparse)
-l lambda: L1 regularization weight (default 0.1)
-c cost: cost of each sample (default 1.0)
-r speed_up_rate: sample 1/r fraction of non-zero features to estimate gradient (default r = ceil(min( 5DK/(Clog(K)nnz(X)), nnz(X)/(5N) )) )
-q split_up_rate: divide all classes into q disjoint subsets (default 1)
-m max_iter: maximum number of iterations allowed if -h not used (default 50)
-u uniform_sampling: use uniform sampling instead of importance sampling (default not)
-g max_select: maximum number of dual variables selected during search (default: -1 (i.e. dynamically adjusted during iterations) )
-p post_train_iter: #iter of post-training without L1R (default auto)
-h <file>: using accuracy on heldout file '<file>' to terminate iterations
-e early_terminate: how many iterations of non-increasing heldout accuracy required to earyly stop (default 3)
Train models on data sets "rcv1_regions" and "sector": $ ./multiTrain -h ./examples/rcv1_regions/rcv1_regions.heldout ./examples/rcv1_regions/rcv1_regions.train rcv1_regions.model $ ./multiTrain -h ./examples/sector/sector/sector.heldout ./examples/sector/sector/sector.train sector.model Note that models generated by post training will be stored separately with name "rcv1_regions.model.p" and "sector.model.p".
$ ./multiPred [testfile] [model] (-p S <output_file>) (k)this command compute top k accuracy and print top S <label>:<prediction score> pairs to <output_file>, one line for each instance. (default S=0 and no file is generated; k = 1)
Compute top-1 accuracy of the model generated: $ ./multiPred ./examples/rcv1_regions/rcv1_regions.test rcv1_regions.model 1 Compute test accuracy of the model generated by post training: $ ./multiPred ./examples/sector/sector.test sector.model.p 1 Compute (top-1) test accuracy and also print top-5 "label:score" pairs to file "sector.table": $ ./multiPred ./examples/sector/sector.test sector.model -p 5 sector.table 1
For data sets used in our paper, we provide a simple script to download, train and test on them automatically. For example, one can run make rcv1_regions to get all above things done for all data sets we used.
We use libsvm format:
<true label 1>,<true label 2>,...,<true label T> <index1>:<value1> <index2>:<value2> ...
.
.
.
$ head -1 ./examples/multiclass/sector/sector.train
53 1:0.00049 2:0.0009 3:4e-05 5:0.00054 6:0.00458 8:0.01302 ... 41303:0.14897
$ head -1 ./examples/multilabel/rcv1_regions.train
53,112 440:0.046331 730:0.098266 ... 46694:0.062828
Our model file format looks like: nr_class <K(number of classes)> label <label 1> ... <label K> nr_features <D(number of features)> <nnz(w[0])> <index 1>:<w[0][index 1]> <index 2>:<w[0][index 2]> ... <index nnz(w[0])>:<w[0][index nnz(w[0])]> <nnz(w[1])> <index 1>:<w[1][index 1]> <index 2>:<w[1][index 2]> ... <index nnz(w[1])>:<w[1][index nnz(w[1])]> . . . <nnz(w[D-1])> ...
$ head -5 model
nr_class 228
label 293 ... 171 352
nr_feature 47237
0
6 65:0.223974 3:0.00958934 17:0.0270412 11:0.134858 42:-0.0397849 98:-0.0188011
PD-Sparse: A Primal and Dual Sparse Approach to Extreme Multiclass and Multilabel Classification.
Ian E.H. Yen*, Xiangru Huang*, Kai Zhong, Pradeep Ravikumar and Inderjit S. Dhillon. (* equally contributed)
In International Conference on Machine Learning (ICML), 2016. [pdf]
For any suggestion and bug report, feel free to contact: Ian En-Hsu Yen(eyan@cs.cmu.edu), Xiangru Huang(xrhuang@cs.utexas.edu)