ModelDB: A System to Manage Machine Learning Models
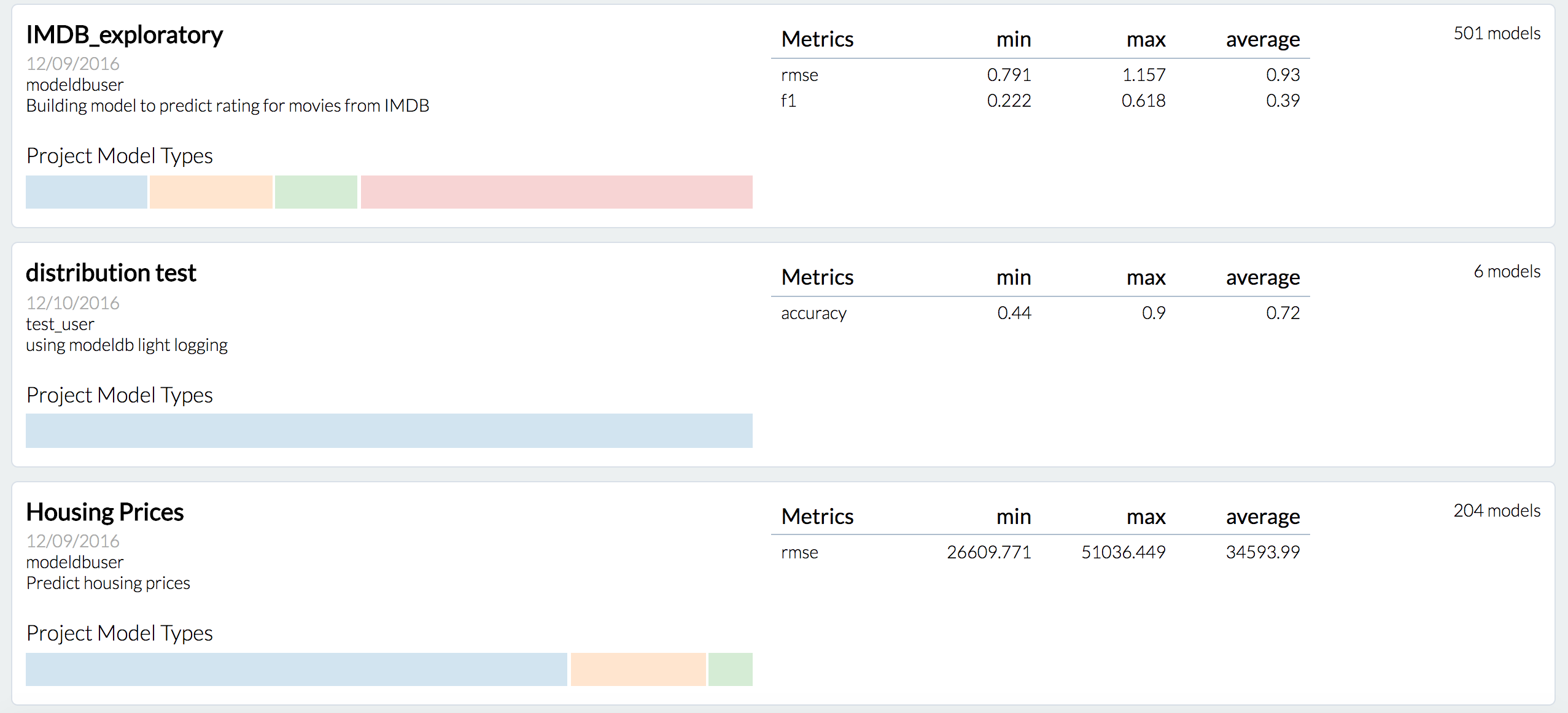
Companies often build hundreds of models a day (e.g., churn, recommendation, credit default). However, there is no practical way to manage all the models that are built over time. This lack of tooling leads to insights being lost, resources wasted on re-generating old results, and difficulty collaborating. ModelDB is an end-to-end system that tracks models as they are built, extracts and stores relevant metadata (e.g., hyperparameters, data sources) for models, and makes this data available for easy querying and visualization.
Publications and Talks:
ModelDB Short paper, HILDA@SIGMOD 2016
Talk at Spark Summit 2016
Full Paper, in preparation
Publications and Talks:
ModelDB Short paper, HILDA@SIGMOD 2016
Talk at Spark Summit 2016
Full Paper, in preparation
SeeDB: A Data-Driven Visualization Recommender System
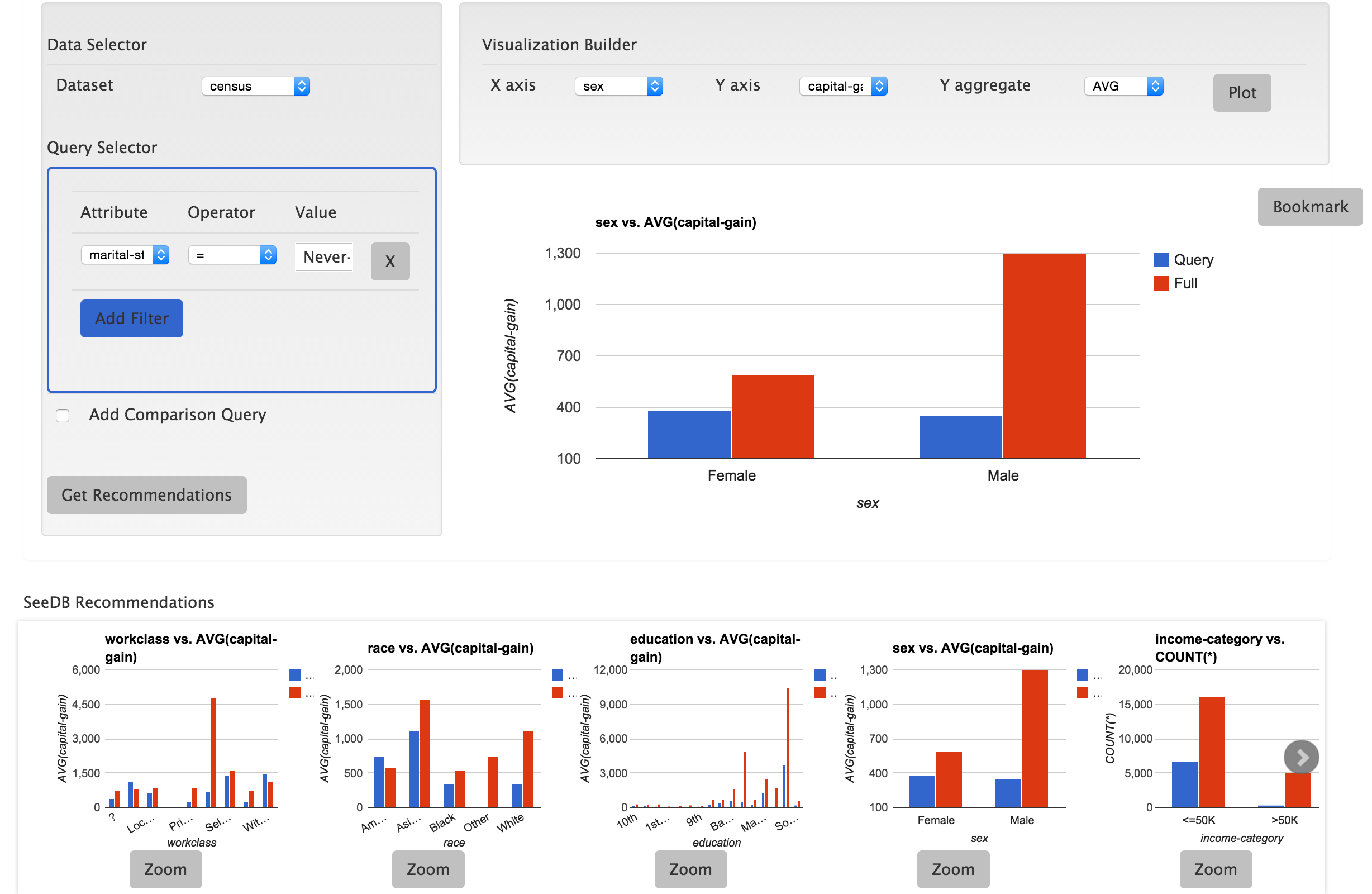
Data analysts often build visualizations as the first step in their analytical workflow. However, when working with high-dimensional datasets, identifying visualizations that show relevant or desired trends in data can be laborious. We propose SEEDB, a visualization recommendation engine to facilitate fast visual analysis: given a subset of data to be studied, SeeDB intelligently explores the space of visualizations, evaluates promising visualizations for trends, and recommends those it deems most “useful” or “interesting”. The two major obstacles in recommending interesting visualizations are (a) scale: evaluating a large number of candidate visualizations while responding within interactive time scales, and (b) utility: identifying an appropriate metric for assessing interestingness of visualizations. For the former, SeeDB introduces pruning optimizations to quickly identify high-utility visualizations and sharing optimizations to maximize sharing of computation across visualizations. For the latter, as a first step, we adopt a deviation-based metric for visualization utility, while indicating how we may be able to generalize it to other factors influencing utility. We implement SeeDB as a middleware layer that can run on top of any DBMS. Our experiments show that our framework can identify interesting visualizations with high accuracy. Our optimizations lead to multiple orders of magnitude speedup on relational row and column stores and provide recommendations at interactive time scales. Finally, we demonstrate via a user study the effectiveness of our deviation-based utility metric and the value of recommendations in supporting visual analytics.
Publications:
SeeDB Demo, VLDB 2014
BigDawg Demo, VLDB 2015
Full Paper, PVLDB Volume 8, Issue 13
Publications:
SeeDB Demo, VLDB 2014
BigDawg Demo, VLDB 2015
Full Paper, PVLDB Volume 8, Issue 13
GenBase: a complex analytics genomics benchmark
This paper introduces a new benchmark designed to test database management system (DBMS) performance on a mix of data management tasks (joins, filters, etc.) and complex analytics (regression, singular value decomposition, etc.) Such mixed workloads are prevalent in a number of application areas including most science workloads and web analytics. As a specific use case, we have chosen genomics data for our benchmark and have constructed a collection of typical tasks in this domain. In addition to being representative of a mixed data management and analytics workload, this benchmark is also meant to scale to large dataset sizes and multiple nodes across a cluster. Besides presenting this benchmark, we have run it on a variety of storage systems including traditional row stores, newer column stores, Hadoop, and an array DBMS. We present performance numbers on all systems on single and multiple nodes, and show that performance differs by orders of magnitude between the various solutions. In addition, we demonstrate that most platforms have scalability issues. We also test offloading the analytics onto a coprocessor. The intent of this benchmark is to focus research interest in this area; to this end, all of our data, data generators, and scripts are available on our web site.
Publications:
Full paper, SIGMOD 2014
Publications:
Full paper, SIGMOD 2014
CHIC: a combination-based recommendation system
Current recommender systems are focused largely on recommending items based on similarity. For instance, Netflix can recommend movies similar to previously viewed movies, and Amazon can recommend items based on ratings of similar users. Although similarity-based recommendation works well for books and movies, it provides an incomplete solution for items such as clothing or furniture which are inherently used in combination with other items of the same type, e.g., shirt with pants, and desk with a chair. As a result, the decision to buy a clothing or furniture item depends not only on the item itself, but also on how well it works with other items of that type. Recommending such items therefore requires a combination-based recommendation system that given an item, can suggest interesting and diverse combinations containing that item. This problem is challenging because features affecting combination quality are often difficult to identify; quality, being a function of all items in the combination, cannot be computed independently; and there are an exponential number of combinations to explore. In this demonstration, we present CHIC, a first-of-its-kind, combination-based recommendation system for clothing. The audience will interact with our system through the CHIC mobile app which allows the user to take a picture of a clothing item and search for interesting combinations containing the item instantly. The audience can also compete with CHIC to create alternate ensembles and compare quality. Finally, we highlight via visualizations the core modules of CHIC including model building and our novel search and classification algorithm, C-Search.
Publications:
CHIC Demo, SIGMOD 2013
Publications:
CHIC Demo, SIGMOD 2013