I recently defended my Ph.D. dissertation, "Machine Learning for Reconstructing Dynamic Protein Structures from Cryo-EM Images". I was part of the Computation and Biology group at the MIT Computer Science and Artificial Intelligence Lab, advised by Bonnie Berger (MIT Mathematics) and Joey Davis (MIT Biology).
Prior to MIT, I was a scientific programmer at D. E. Shaw Research, where I developed algorithms and infrastructure for predicting protein-small molecule binding free energies via molecular dynamics simulations. I graduated from the University of Virginia, where I worked with Michael Shirts on studying the thermodynamics of protein folding with Hamiltonian Monte Carlo simulations.
For more information, see my curriculum vitae.
Proteins and protein complexes are dynamic molecular machines that undergo large structural changes in order to function. Looking beyond static 3D protein structures, how can we study distributions of protein conformations that are relevant to function (or dysfunction)?
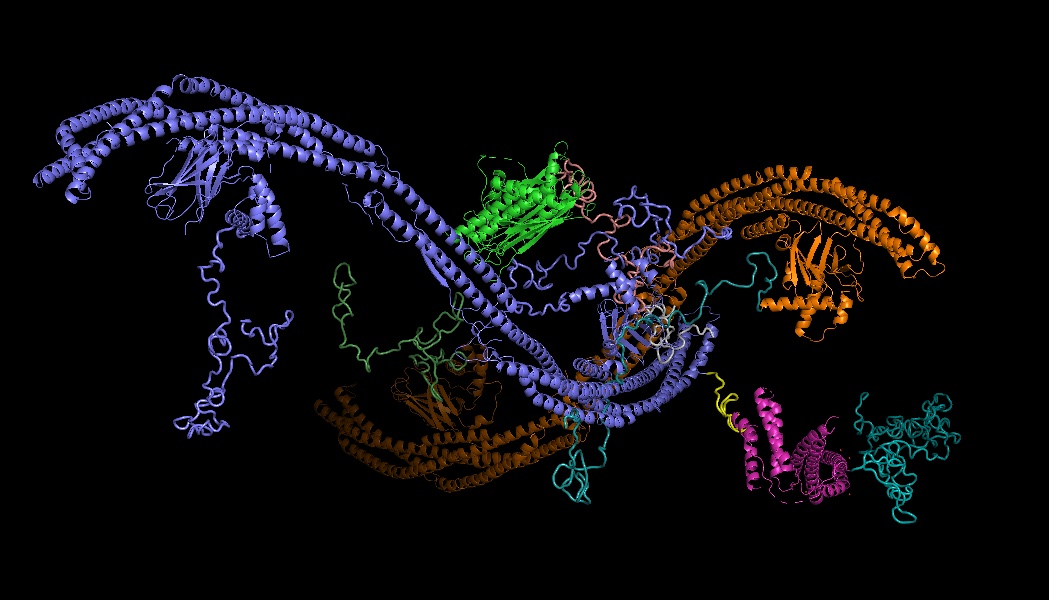
The primary focus of my PhD work is to develop neural methods for reconstructing distributions of 3D protein structure from 2D cryo-electron microscopy (cryo-EM) image data. Cryo-EM is an experimental technique for protein structure determination that has enourmous potential for visualizing the details of molecular motions at (near-)atomic resolution. However, protein structural variability severely complicates the computational 3D reconstruction task, the so-called "heterogeneity problem".
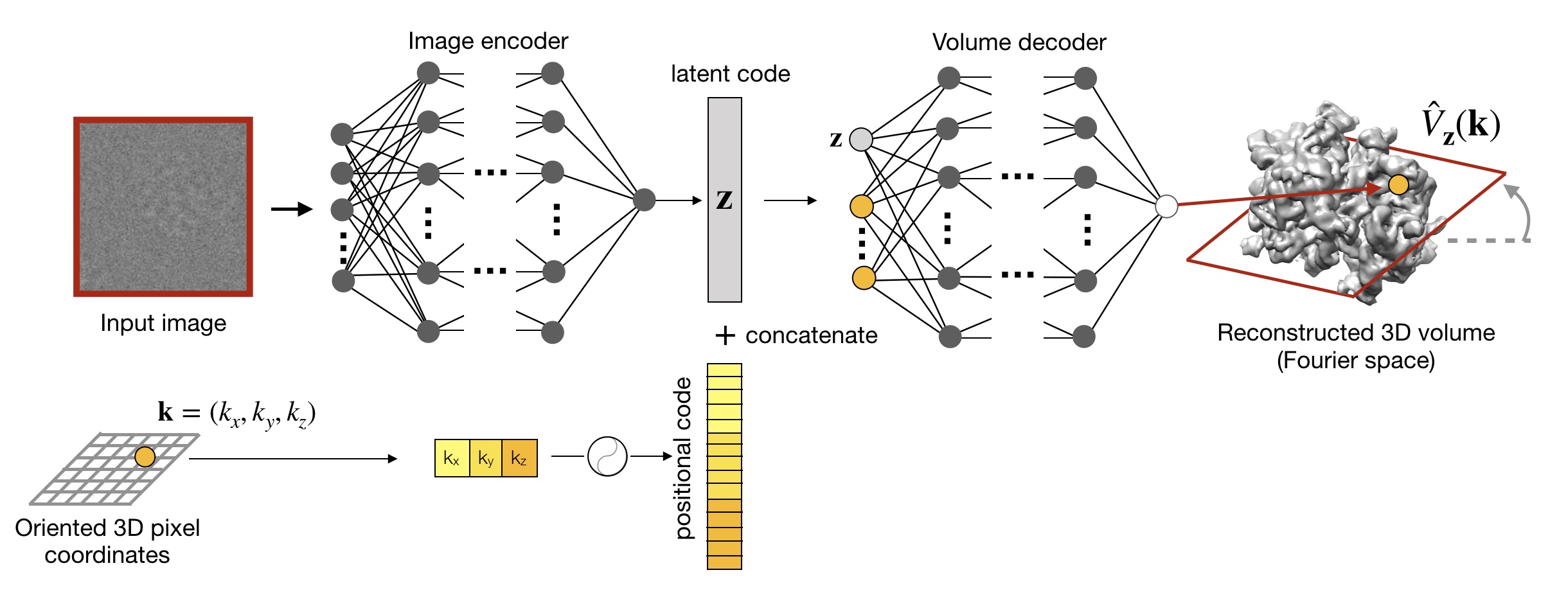
My Ph.D. has focused on developing new deep learning methods for heterogeneous cryo-EM reconstruction to help realize this opportunity to study dynamic protein structures. See below for publications and presentations describing our work:
Research into improvements to cryoDRGN and related algorithms is actively ongoing. Please reach out if you'd like to learn more!
During the covid-19 pandemic, we formulated a new language modeling task for predicting viral escape mutations:
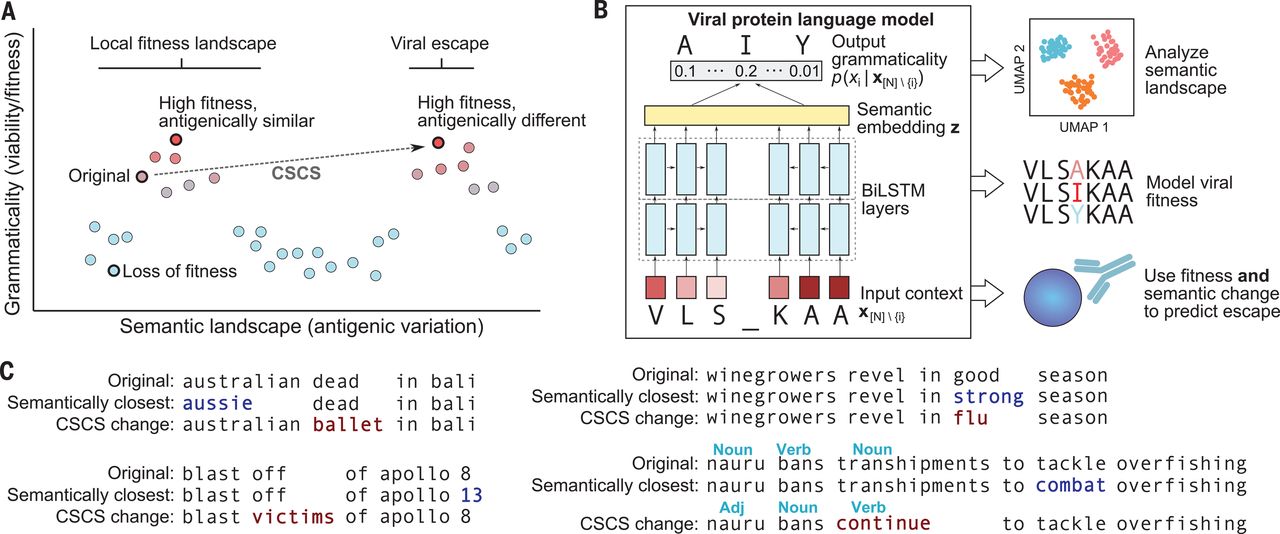
Learning the language of viral evolution and escape
Brian Hie, Ellen D. Zhong, Bonnie Berger, and Bryan Bryson.
Science, January 2021.
During a rotation in the Boyden lab, I developed simple statistical models for inferring temporal dynamics of gene expression from RNA timestamps:
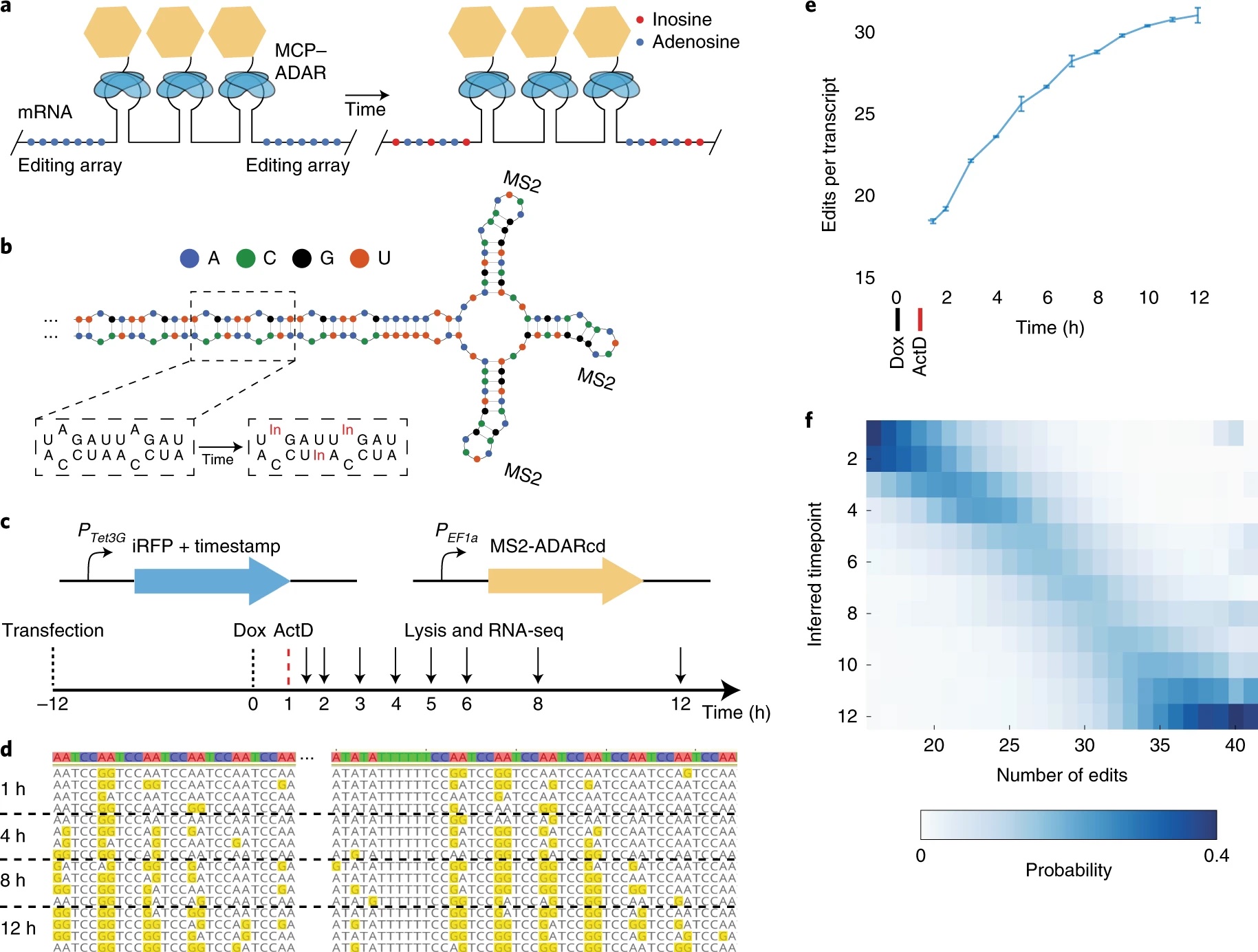
RNA timestamps identify the age of single molecules in RNA sequencing
Sam Rodriques, Linlin Chen, Sonya Liu, Ellen D. Zhong, Joe Scherrer, Ed Boyden, and Fei Chen.
Nature Biotechnology, October 2020.
In my free time, I enjoy rock-climbing, exploring the outdoors, and cooking elaborate meals.