 |
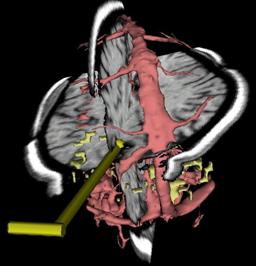
| D. Gering, A. Nabavi, R. Kikinis, N. Hata, L. Odonnell, W. Eric L. Grimson, F. Jolesz, P. Black, W. Wells III.
An Integrated Visualization System for Surgical Planning and Guidance Using Image Fusion and an Open MR.
Journal of Magnetic Resonance Imaging, Vol 13, pp. 967-975, June, 2001.
|
| ABSTRACT: A surgical guidance and visualization system is presented, which uniquely integrates capabilities for data
analysis and on-line interventional guidance into the setting of interventional MRI. Various pre-operative scans
(T1- and T2-weighted MRI, MR angiography, and functional MRI (fMRI)) are fused and automatically aligned with the
operating field of the interventional MR system. Both pre-surgical and intra-operative data may be segmented to
generate three-dimensional surface models of key anatomical and functional structures. Models are combined in a
three-dimensional scene along with reformatted slices that are driven by a tracked surgical device. Thus,
pre-operative data augments interventional imaging to expedite tissue characterization and precise localization and
targeting. As the surgery progresses, and anatomical changes subsequently reduce the relevance of preoperative data,
interventional data is refreshed for software navigation in true real time. The system has been applied in 45
neurosurgical cases and found to have beneficial utility for planning and guidance. |
|
 | | 523K |
|
|
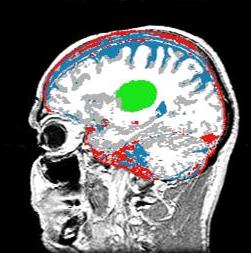 |
| D. Gering.
Recognizing Deviations from Normalcy for Brain Tumor Segmentation.
MIT Ph.D. Thesis, June, 2003.
|
| ABSTRACT: A framework is proposed for the segmentation of brain tumors from MRI. Instead of training on pathology,
the proposed method trains exclusively on healthy tissue. The algorithm attempts to recognize deviations
from normalcy in order to compute a fitness map over the image associated with the presence of pathology.
The resulting fitness map may then be used by conventional image segmentation techniques for honing in on
boundary delineation. Such an approach is applicable to structures that are too irregular, in both shape
and texture, to permit construction of comprehensive training sets.
We develop the method of diagonalized nearest neighbor pattern recognition, and we use it to demonstrate that
recognizing deviations from normalcy requires a rich understanding of context. Therefore, we propose a framework
for a Contextual Dependency Network (CDN) that incorporates context at multiple levels: voxel intensities,
neighborhood coherence, intra-structure properties, inter-structure relationships, and user input.
Information flows bi-directionally between the layers via multi-level Markov random fields or iterated Bayesian classification.
A simple instantiation of the framework has been implemented to perform preliminary experiments on synthetic and MRI data. |
|
|
|
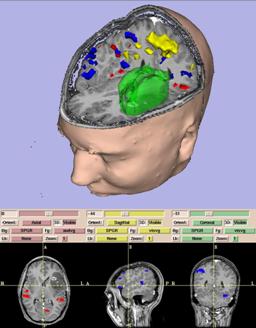 |
| D. Gering, A. Nabavi, R. Kikinis, W. Eric L. Grimson, N. Hata, P. Everett, F. Jolesz, W. Wells III.
An Integrated Visualization System for Surgical Planning and Guidance using Image Fusion and Interventional Imaging.
Medical Image Computing and Computer-Assisted Intervention (MICCAI), Cambridge England, pp. 809-819, Sept 1999.
Springer-Verlag
|
| ABSTRACT: We present a software package which uniquely integrates several facets of image-guided medicine into a single portable,
extendable environment. It provides capabilities for automatic registration, semiautomatic segmentation, 3D surface model
generation, 3D visualization, and quantitative analysis of various medical scans. We describe its system architecture,
wide range of applications, and novel integration with an interventional Magnetic Resonance (MR) scanner to augment
intraoperative imaging with pre-operative data. Analysis previously reserved for pre-operative data can now be applied to
exploring the anatomical changes as the surgery progresses. Surgical instruments are tracked and used to drive the location
of reformatted slices. Real-time scans are visualized as slices in the same 3D view along with the pre-operative slices and
surface models. The system has been applied in over 20 neurosurgical cases at Brigham and Women's Hospital, and continues to
be routinely used for 1-3 cases per week. |
|
 | | 343K |
|
|
 |
| D. Gering, W. Wells.
Object Modeling using Tomography and Photography.
IEEE Workshop on Multi-View Modeling and Analysis of Visual Scenes (in conjuction with CVPR), Fort Collins, CO, Jun 26, 1999, pp. 11-18.
|
| ABSTRACT: This paper explores techniques for constructing a 3D computer model of an object from the real world by
applying tomographic methods to a sequence of photographic images. While some existing methods can better
handle occlusion and concavities, the techniques proposed here have the advantageous capability of generating
very high-resolution models with attractive speed and simplicity. The application of these methods is presently
limited to an appropriate class of mostly convex objects with Lambertian surfaces. The results are volume rendered
or surface rendered to produce an interactive display of the object with near life-like realism. |
|
 | | 504K |
|
|
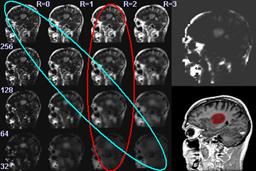 |
| D. Gering.
Diagonalized Nearest Neighbor Pattern Recognition for Brain Tumor Segmentation.
Medical Image Computing and Computer-Assisted Intervention (MICCAI), Montreal Canada, pp II-670, Nov 2003.
Springer-Verlag
|
| ABSTRACT: A new method is proposed for automatic recognition of brain tumors from MRI. The prevailing convention in the
literature has been for humans to perform the recognition component of tumor segmentation, while computers
automatically compute boundary delineation. This concept manifests as clinical tools where the user is required to
select seed points or draw initial contours. The goal of this paper is to experiment with automating the recognition
component of the image segmentation process. The main idea is to compute a map of the probability of pathology, and
then segment this map instead of the original input intensity image. Alternatively, the map could be used as a feature
channel in an existing tumor segmentation method. We compute our map by performing nearest neighbor pattern matching
modified with our novel method of 'diagonalization'. Results are presented for a publicly available data set of brain tumors. |
|
 | | 427K |
|
|
 |
| D. Gering, W. Eric L. Grimson, R. Kikinis.
Recognizing Deviations from Normalcy for Brain Tumor Segmentation.
Medical Image Computing and Computer-Assisted Intervention (MICCAI), Tokyo, Japan, pp. 388-395, Sept 2002.
Springer-Verlag
|
| ABSTRACT: A framework is proposed for the segmentation of brain tumors from MRI. Instead of training on pathology,
the proposed method trains exclusively on healthy tissue. The algorithm attempts to recognize deviations from
normalcy in order to compute a fitness map over the image associated with the presence of pathology. The
resulting fitness map may then be used by conventional image segmentation techniques for honing in on boundary
delineation. Such an approach is applicable to structures that are too irregular, in both shape and texture,
to permit construction of comprehensive training sets. The technique is an extension of EM segmentation that
considers information on five layers: voxel intensities, neighborhood coherence, intra-structure properties,
inter-structure relationships, and user input. Information flows between the layers via multi-level Markov random
fields and Bayesian classification. A simple instantiation of the framework has been implemented to perform
preliminary experiments on synthetic and MRI data. |
|
 | | 96K |
|
|
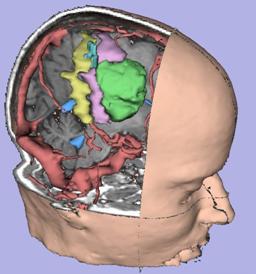 |
| D. Gering.
A System for Surgical Planning and Guidance using Image Fusion and Interventional MR.
MIT Master's Thesis, 1999.
|
| ABSTRACT: In this thesis, we present a computerized surgical assistant whose core functionality is embodied
in a software package we call the 3D Slicer. We describe its system architecture and its novel integration
with an interventional Magnetic Resonance (MR) scanner. We discuss the 3D Slicer's wide range of applications
including guiding biopsies and craniotomies in the operating room, offering diagnostic visualization and
surgical planning in the clinic, and facilitating research into brain shift and volumetric studies in the lab.
The 3D Slicer uniquely integrates several facets of image-guided medicine into a single environment. It provides
capabilities for automatic registration, semi-automatic segmentation, surface model generation, 3D visualization,
and quantitative analysis of various medical scans.
We formed the first system to augment intra-operative imaging performed with an open MR scanner with this full array
of pre-operative data. The same analysis previously reserved for pre-operative data can now be applied to exploring
the anatomical changes as the surgery progresses. Surgical instruments are tracked and used to drive the location
of reformatted slices. Real-time scans are visualized as slices in the same 3D view along with the pre-operative
slices and surface models. The system has been applied in over 20 neurosurgical cases at Brigham and Women's Hospital,
and continues to be routinely used for 1-2 cases per week. |
|
 | | 9.5M |
|
|
Links
|
|