
Segger - Aligning structures to regions
- Example: GroEL
- Fitting options
- GroeL example: aligning the structure to multiple, single regions
- Example: Bacteriophage lambda
1.1 Segmenting the map
First, the density map must be segmented. Here we will use as an example the density map of GroEL at 4.2Å resolution. This map can be download from this link. Open the map, and open the Segger dialog as described on the segmentation page. Select a threshold of 0.9, and in the Segmenting options, enter 4 steps of size 8, and then press Segment. The result will be 14 regions, with each region corresponding to a single protein. The density map and segmented regions are shown below.
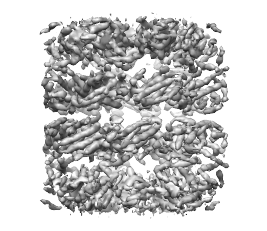

Side note: the segmentation threshold does not actually influence the number of regions obtained. At other thresholds (at which the background noise is not included), the same number of regions can be obtained by the smoothing and grouping method.
Side note: For this example we used large smoothing steps intentionally. When taking smaller steps, less accurate segmentation regions were obtained. This is likely due to noise in the density map. Applying more smoothing in the first step is helping to suppress this noise.
The structure of a single protein in GroEL can be obtained from PDB:1xck. After downloading this structure, it should be placed in the same directory as the segmented GroEL density map (emd_5001.mrc). It can then be opened using the Segger interface, by clicking on the drop-down menu to the right of the 'Structure to fit' label. This drop-down menu shows all PDB files in the same dirctory as the selected density map (which should be emd_5001.mrc). Selecting 1XCK.pdb will open the structure.
Select a single chain of this structure, e.g. chain A, and save it to its own PDB file, e.g. 1xck_A.pdb. Then use the drop-down menu to the right of 'Structure to fit' label to select and open this structure by itself. Alternatively, you can delete all chains other than A, and then choose 1xck.pdb in the drop-down menu.
Once the structure of a single protein is selected in the drop-down menu to the right of the Structure to fit label, its principal axes can be shown by selecting Fitting at the top of the Segger dialog, and then selecting Show molecule axes. The structure and axes are shown in the image below.

1.3 Fitting the structure by aligning it to a region
We can now align this structure to a segmented region in the density map. One way to do this is using the principal-axes transform. To see how this would work, select a single segmented region, and then select Regions->Show only selected, to show this region alone, then Regions->Make transparent to allow us to see through the region surface, and then Regions->Show axes for selected to show the principal axes for this region. The resulting view is shown in the image below.

By looking at the two images above, shown again below side by side, it can be seen that principal axes are roughly the same with respect to the structure and to the region. The structure can thus be aligned to the region by matching the centers and the principal axes of the structure to those of the region.


To perform the alignment, make sure the structure is selected in the field to the right of 'Structure to fit' label, one of the regions is selected, and then press the Fit button at the bottom of the Segger dialog. In a few seconds, the structure will have been moved to fit right into the region, as shown below:

Side note: Showing the principal axes of the structure or the region is not required to complete the alignment process. They are only shown here for illustrative purposes.
Side note: The aligment process here only achieves a rigid fit. This assumes the structure of the molecule being fit should be the same in both the cryo-EM and crystallographic states for a good fit to be obtained. The latter may not always be true, for example some proteins may have different conformations under different conditions. In such cases, a flexible-fitting method, such as Direx or MDFF should be used.
To see more fitting options, press the Options button at the bottom of the Segger window (unless you've already pressed it). The fitting options are shown below:

2.1 Parameters for density map
During the fitting process, a density map is generated for the structure. This density map is used to compute a cross-correlations score for the fit. After the structure is aligned to one or more region, the structure and density map are moved so as to increase the cross-correlation score, resulting in better fits.
The default density map resolution and grid spacing are taken from the density map selected in the field to the right of 'Map to segment'. You can enter different resolutions and grid spacing.
2.2 Which regions to align the structure to
There are several possibilities for aligning structures to regions:
Combined selected regions: This is the default, and in this mode, the structure is aligned to the selected region. If more than one regions are selected, the structure is aligned to the combined regions.
Each selected region: The structure is aligned to each selected region. This means that more than one alignment will take place, and multiple fits will result. To see each fit, make sure to sheck the button to the left of 'Save each fit when doing multiple alignments'. Note that if no regions are selected, and this mode is chosen, then the structure is aligned to each region in the current segmentation.
Groups of regions including selected region: If you're not sure which group of regions will create the best alignment and thus the best fit, but you have an idea of which region might be involved in this group, select that region, and select this mode. When you press the Fit button, groups of regions will be automatically generated, including the selected region. The structure will be aligned to each group, and the best resulting fit will be displayed at the end.
Groups of regions including all regions: If you're not sure which group of regions will create the best alignment and thus the best fit, and don't know that any region in particular is part of this group of regions, and select this mode. When you press the Fit button, groups of regions will be automatically generated, including the all existing regions. The structure will be aligned to each group, and the best resulting fit will be displayed at the end. Note that if you start with many regions, many groups will be generated, and this process may take a while. After it is done, all the fits are recorded and saved. To see the best fits, enter the number of fits you would like to see to the right of 'Top number of fits to place', such as 1, 7, etc., and then press the Place button. The fits with the highest cross-correlations scores will be shown, and the structure will be duplicated and saved in each fit position.
2.3 Alignment method
Align by principal axes: This alignment method is described above in the example, and is fast. However it may not always find the right fit. It is likely to fail for structures that are symmetric (sphere or rod-like). Note that you don't have to show the axes of either the structure or the molecule to use this method, as in the exampe above, that was only for illustration purposes.
Rotational search:
Use this method when the principal axes method fails. It involves aligning the centers of the structure and region, groups of regions (as indicated by teh choice under 'Which regions to align the structure to', and then searching only through different orientations.
2.4 Saved fitted structures
The name of each saved fitted structure will be struc_name_f#.pdb, where struc_name is the name of the original PDB file the structure came from, and # is 1..n, where n is the number of fitted structures saved.
Back to the GroEL example, make sure no regions are selected, and then check the 'Save each fit when doing multiple alignments' button. Also choose the 'Each selected region' under 'Which regions to align the structure to', and press the Fit button. Under this mode, the structure is aligned to each selected region. But if no regions are selected, then the structure is aliged to each region.
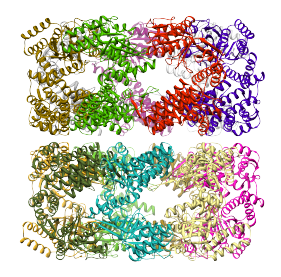
Since in this example, each region corresponds to a single protein, the structure is aligned successfuly to each one, reproducing the strcture of the entire complex, as shown below:
2.1 Getting and segmenting the density map
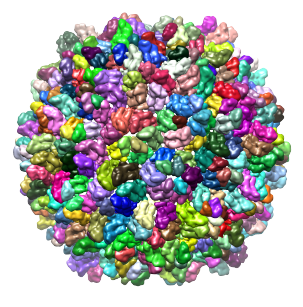
As another example, we will use the density map of bacteriophage lambda at 14.5Å resolution, which can be downloaded from this link. After opening the map, adjust its threshold to 3.5, and select it in the field to the right of 'Segment map'. Enter 1 step of size 10.0 in the Segmenting Options, and press Segment. Because this map is very large, taking more steps typically results in memory problems on 32-bit systems. A total of 394 regions result. The density map and the segmented regions are shown below.

2.2 Choosing regions to keep
The regions that make up an asymmetric unit (ASU) can be seen in this segmentation, and can be selected together within the Chimera window - use Ctrl+Click for the first region and Shift+Ctrl+Click for all subsequent regions. Then, to keep only these regions, choose Regions->Invert selection, and then Regions->Delete selected. The remaining regions are shown in the image below.

As can be seen above, there are 10 regions, however an ASU actually contains only 7 proteins. Thus, for some of the proteins, two regions correspond to each protein. If we tried to smooth further, to attempt to obtain a single region for each protein, regions from different proteins merge first. So for this map, when smoothing, a point can't be reached in which each region corresponds to a protein. This leads to having to deal with over-smoothing and/or multiple regions per protein.
2.3 Dealing with over-smoothing and multiple regions per protein
The smoothing and grouping process would ideally be stopped just before regions corresponding to different proteins or molecular components merge. This can only be done by visual inspection of the regions after grouping. If too much smoothing is done, and too few regions are obtained, an intermediate files can be selected in the menu-button to the right of the Regions button, which contains more regions (see the documentation on segmenting a density map). When more than one region corresponds to each protein, to get regions corresponding to single proteins, the regions that belong to the same protein can then be selected, and joined by selecting Regions->Group selected.
2.4 Aligning a structure to regions - user selection
In this example, we can align a protein structure to single and multiple regions. A structure that fits well into this map is PDB:3BQW.
After downloading this file, place it in the same directory as the density map, and select it in the field to the right of 'Structure to fit' label.
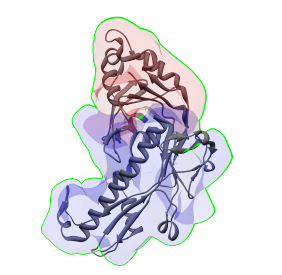
Then choose 'Combined selected regions' under 'Which regions to align the structure to', select one or two regions that appear to correspond to this structure, and press the Fit button. If the right regions are selected, the alignments will look like in the images below. In the image on the left, the structure was aligned to a single region, and on the right, it was aligned to two regions.
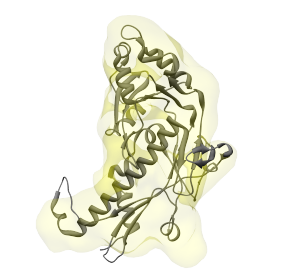
Instead of relying on the you to select the correct regions for alignment, Segger can automatically generate candidate groups of regions to align the structure to. After aligning the structure to each of the generated groups, the alignment with the highest cross-correlation is kept. Groups can be generated in two ways:
2.5 Aligning a structure to regions - automatic grouping starting with one region
In the first way, select a single region, and choose 'Groups of regions including selected region' under 'Which regions to align the structure to'. Then press Fit. In this mode, about 5 groups should be generated, and after trying to align the structure to each one, the fit with the highest cross-correlations is kept.
2.6 Aligning a structure to regions - automatic grouping of all regions
In the second way, choose 'Groups of regions including selected region' under 'Which regions to align the structure to'. Then press Fit.
Groups are generated in combinatorial fashion, however all groups have only adjacent regions (i.e. in every group, every region is adjacent to at least one other region). The latter condition helps to keep the number of groups down dramatically.
The groups are also filtered such that they don't have a volume that is too different than that of the structure (computed from the density map of the structure at the currently selected threshold), and they don't have a bounding radius that is very different than that of the structure. This filtering also helps to decrease the number of groups considered, without eliminating groups that will lead to the correct fit.
Note: When working with a lot of regions, this process can result in very many groups, and the alignment process can take a long time.
In this example, only 22 groups are generated, and the structure is aligned to each of these groups. Both the principal axes method or rotational search produce the same result, however the principal axes method is much faster. When the principal axes method doesn't seem to produce the right result, rotational search should be used instead.
During the alignment process, a file is created which contains all the cross-correlations scores, sorted in decreasing order. This file may show less than 22 fits, since only the fits with unique transforms are kept (different alignments can result in the same final transform for the fitted structure). The file in the same folder as the density map, emd_1507.mrc, and is named emd_1507_fits_sorted.txt. The first 10 entries in the file are:
2 - structure: 3BQW.pdb, cross-correlation: 0.458
3 - structure: 3BQW.pdb, cross-correlation: 0.450
4 - structure: 3BQW.pdb, cross-correlation: 0.447
5 - structure: 3BQW.pdb, cross-correlation: 0.446
6 - structure: 3BQW.pdb, cross-correlation: 0.420
7 - structure: 3BQW.pdb, cross-correlation: 0.409
8 - structure: 3BQW.pdb, cross-correlation: 0.185
9 - structure: 3BQW.pdb, cross-correlation: 0.166
10 - structure: 3BQW.pdb, cross-correlation: 0.163
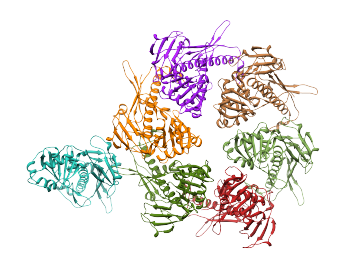
The top 7 cross-correlation scores are much higher than the rest. This indicates that there are 7 good alignments for the structure. In fact, the top 7 alignments produced the correct fits for the protein structure which recreate the entire ASU. To generate the structure of the complete ASU, enter '7' in the field to the right of "Top number of fits to place", and press the Place button. This places the structure in the alignments with the top 7 cross-correlation scores, making a copy of the structure in each alignment, and also saving a .pdb file for each alignment. The results are shown in the image below.
Please contact Greg Pintilie by email with comments and suggestions.

