Research Interests
In my PhD, the focus of my research was on machine learning and medical imaging and recently I have become interested in bioinformatics and in particular its conjunction with medical imaging. The objective of my research is to simultaneously exploit wealth of information in medical image and human genomics to improve understanding of human disease via building statistical and computational methods. Here is a list of my recent manuscripts (see My CV for a complete list):
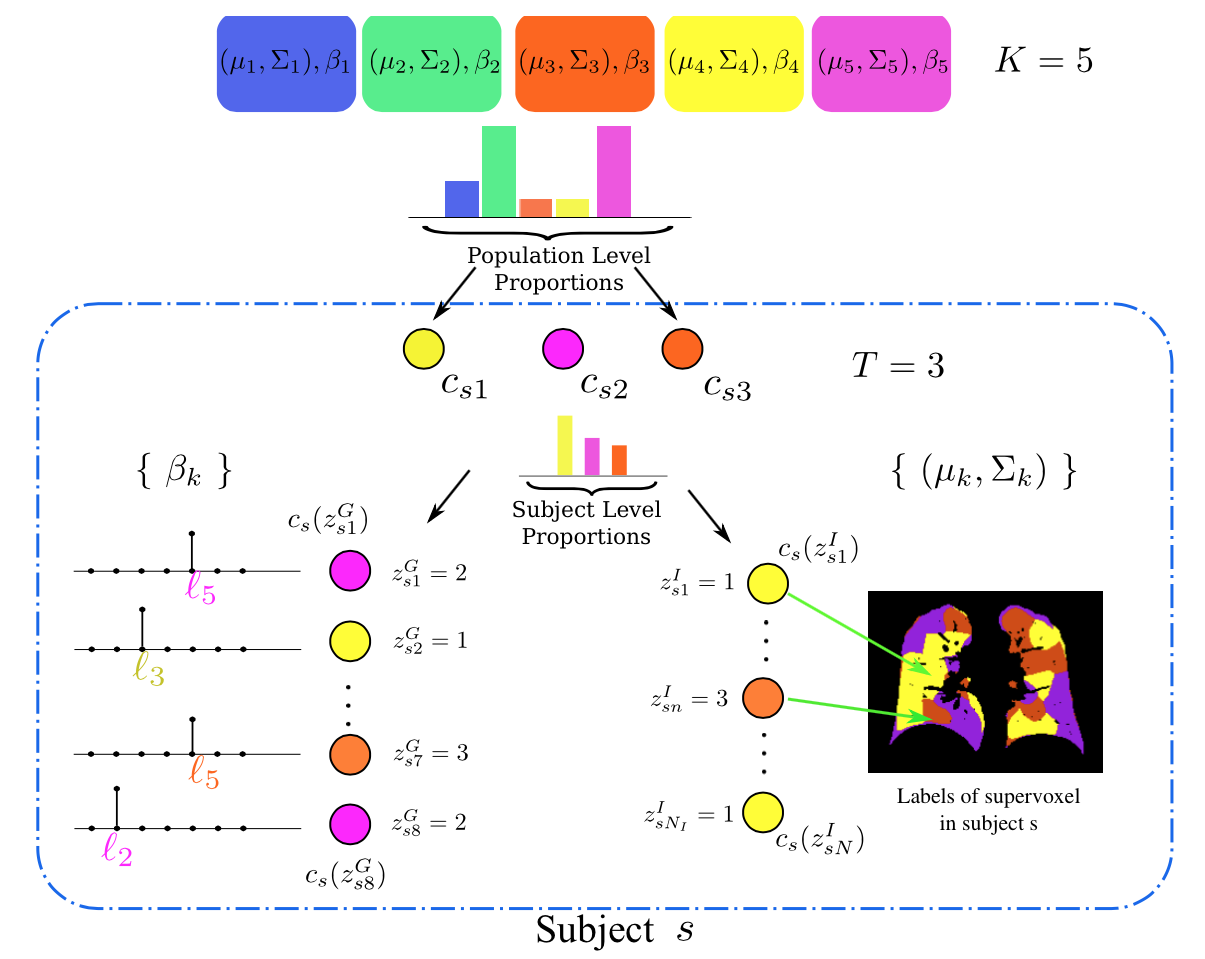
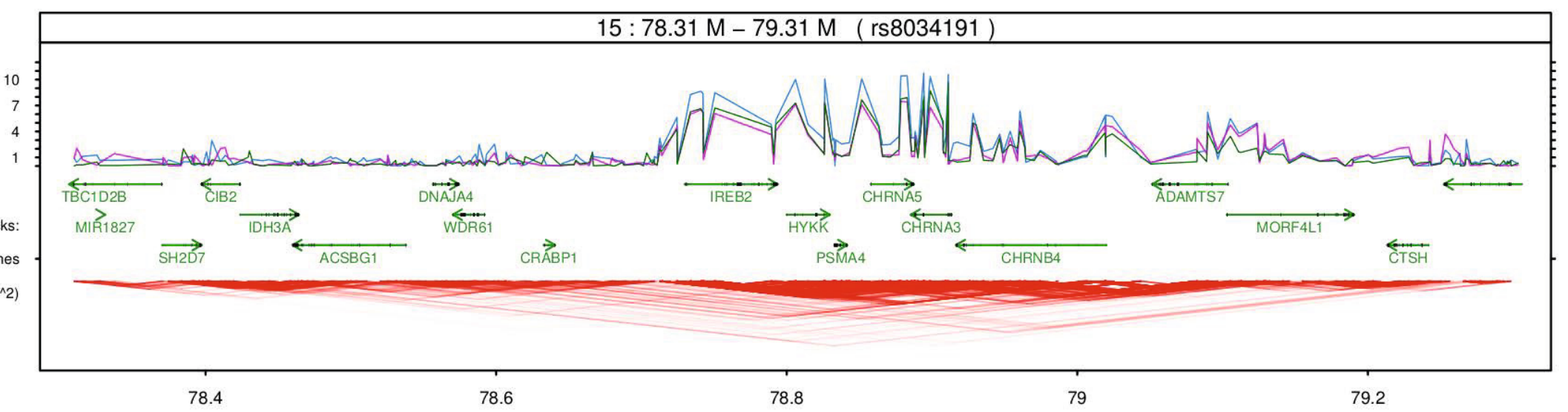
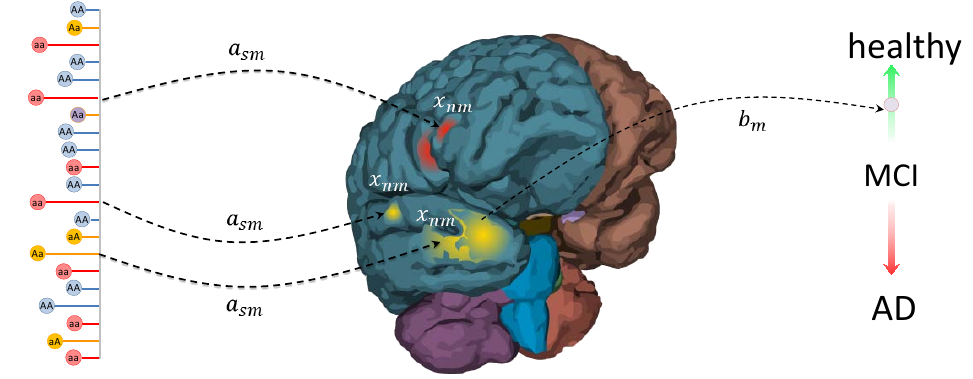
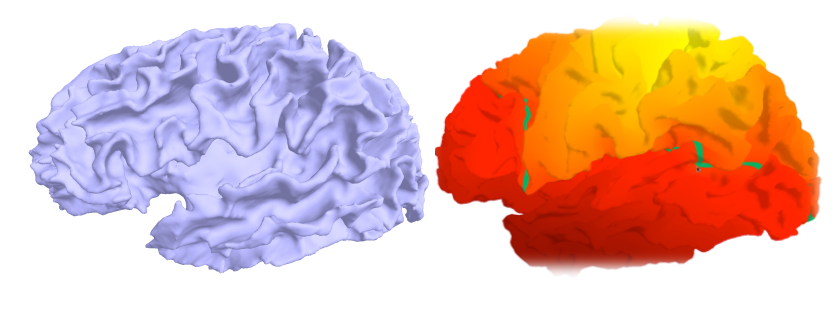
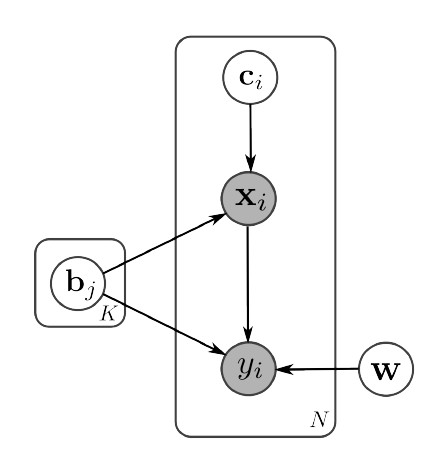
Joint Modeling of Imaging-Genetics
There are two main purposes for combining imaging and genetic information: 1) using imaging as intermediate information (so-called intermediate phenotype) to understand underlying biological processes of a disease, 2) using known genetic markers of the disease to interpret observed anatomy in the imaging data. Here are some of my published works toward those achieving goals:
- K.N. Batmanghelich, A. Dalca, G. Quon, M. Sabuncu, P. Golland. Probabilistic Modeling of Imaging, Genetics and the Diagnosis, IEEE Trans Med Imaging, 35(7):1765-79, 2016.
- K.N. Batmanghelich*, A. Saeedi*, M. Cho, R. Jose, P. Golland. Generative Method to Discover Genetically Driven Image Biomarkers, In Proc. IPMI: International Conference on Information Processing and Medical Imaging, accepted, 2015. (Oral Presentation)
- K.N. Batmanghelich, M. Cho, R. Jose, P. Golland. Spherical Topic Models for Imaging Phenotype Discovery in Genetic Studies, In Proc. BAMBI: Workshop on Bayesian and Graphical Models for Biomedical imaging, International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI), LNCS 8677, pp. 107-117, 2014. (Oral Presentation)
- K.N. Batmanghelich, A.V. Dalca, M.R. Sabuncu, P. Golland. Joint Modeling of Imaging and Genetics, In Proc. IPMI: International Conference on Information Processing and Medical Imaging, LNCS 7917, pp. 766-777, 2013. (Oral Presentation)
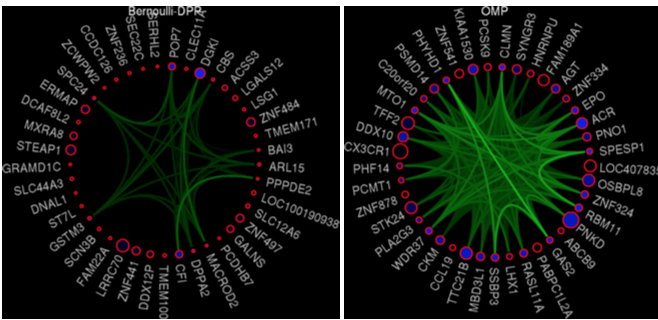
Point Process for Diverse Feature Selection
This work is related to the previous projects. The goal is to find a diverse subset of features that are optimal for a task (e.g. classification or regression) while being diverse with respect to some given side information. We formulate the problem as learning the quality scores of items in a Determinantal Point Process (DPP).
- K.N. Batmanghelich, G. Quon, A. Kuleza, M. Kellis, P. Golland, L. Bornn. Diversifying Sparsity Using Variational Determinantal Point Processes, In Proc. ArXiv , 2014.
Dimensionality Reduction for Medical Imaging Applications
This project addresses the large dimension, low sample size problems (p>>n) in the medical imaging applications and more specifically for the diagnosis of the Alzheimer's disease. The goal is to transform very high-dimensional input to a low-dimensional representation that preserves discriminative signal and the resultant representation is clinically interpretable. We formulate the task as a constrained optimization problem that combines generative and discriminative objectives. We propose a novel large-scale algorithm to solve the resulting optimization problem.
- C. Wachinger, K. Batmanghelich, P. Golland, M. Reuter BrainPrint in the Computer-Aided Diagnosis of Alzheimer's Disease . , Challenge on Computer-Aided Diagnosis of Dementia, MICCAI, 2014. (Second prize in classification challenge)
- N. Batmanghelich, B. Taskar, C. Davatzikos, Generative-Discriminative Basis Learning for Medical Imaging. , IEEE Trans Med Imaging. 2012 Jan;31(1):51-69.
- N. Batmanghelich, B. Taskar, C. Davatzikos, Regularized Tensor Factorization for Multi-Modality Medical Image Classification. , International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI 2011), Toronto, Canada
- N. Batmanghelich, D. Ye, K. Pohl, B. Taskar, C. Davatzikos, Disease Classification and Prediction via Semi-Supervised Dimensionality Reduction, ISBI 2011 (Oral Presentation)
- N. Batmanghelich, B. Taskar, C. Davatzikos, A General and Unifying Framework for Feature Construction. in Image-Based Pattern Classification, 20'th International Conference on Information Processing in Medical Imaging (IPMI), LNCS '09. pp. 423-434, 2009. (Oral Presentation)
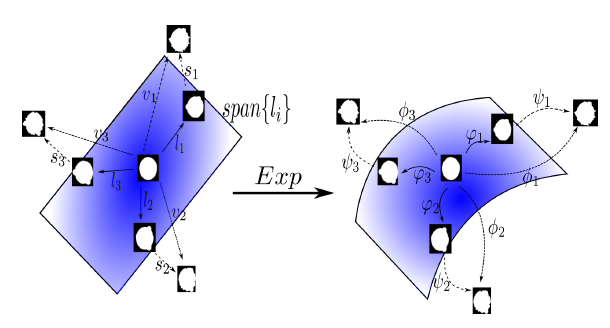
Matrix Decomposition for Deformation Modeling and its applications in Computational Anatomy
Given a set of normal and abnormal brain images, in this project we try to solve an inverse problem: how to decompose a brain deformation to a normal and abnormal deformations from an average brain. The deformations are parameterized by stationary velocity fields and the velocity fields are decomposed to low-rank and sparse components. The algorithm iterates between image registration and the decomposition step and hence can be viewed as a group-wise registration algorithm.
- N. Batmanghelich, A. Gooya, B. Taskar, C. Davatzikos, Application of Trace-Norm and Low-Rank Matrix Decomposition for Computational Anatomy, IEEE Computer Society Workshop on Mathematical Methods in Biomedical Image Analysis (MMBIA10), CVPR 2010.