Highlights
Preprints

|
Autograd: reverse-mode differentiation of native Python
Autograd automatically differentiates native Python and Numpy code. It can handle loops, ifs, recursion and closures, and it can even take derivatives of its own derivatives. It uses reverse-mode differentiation (a.k.a. backpropagation), which means it's efficient for gradient-based optimization. Check out the tutorial and the examples directory. Dougal Maclaurin, David Duvenaud, Matthew Johnsoncode | bibtex |
Publications
|
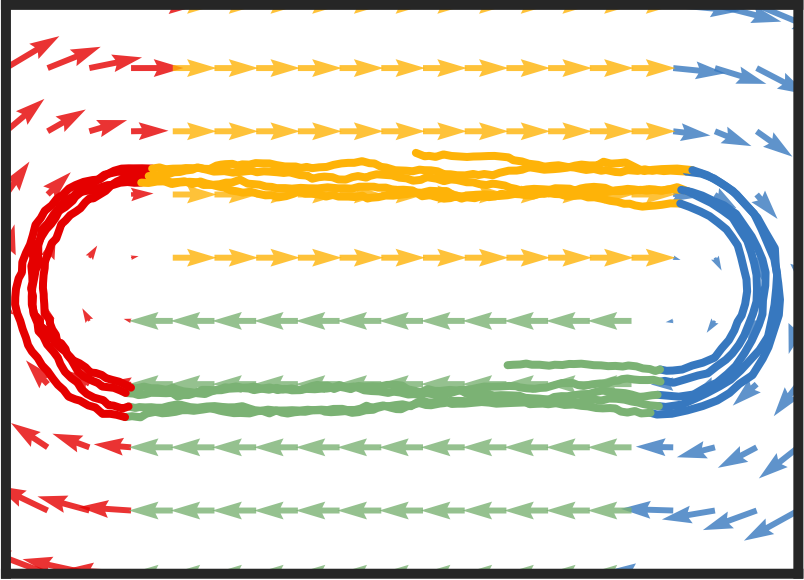
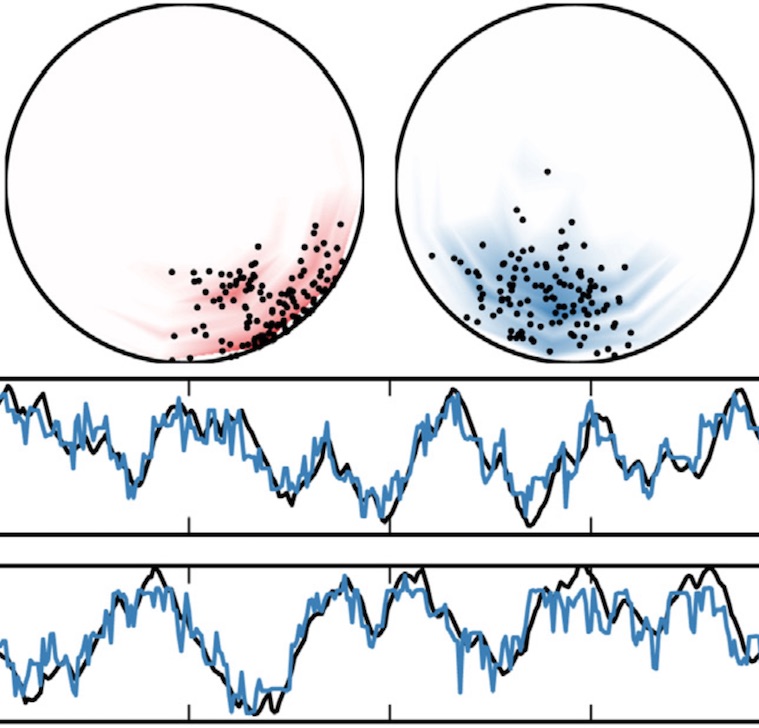
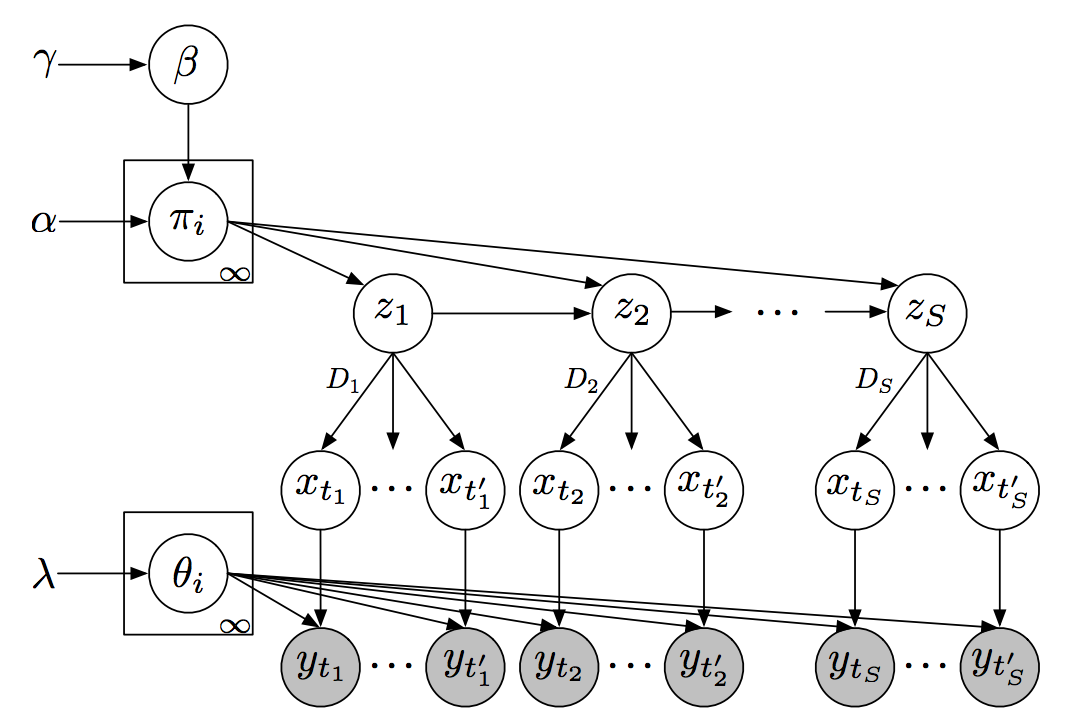
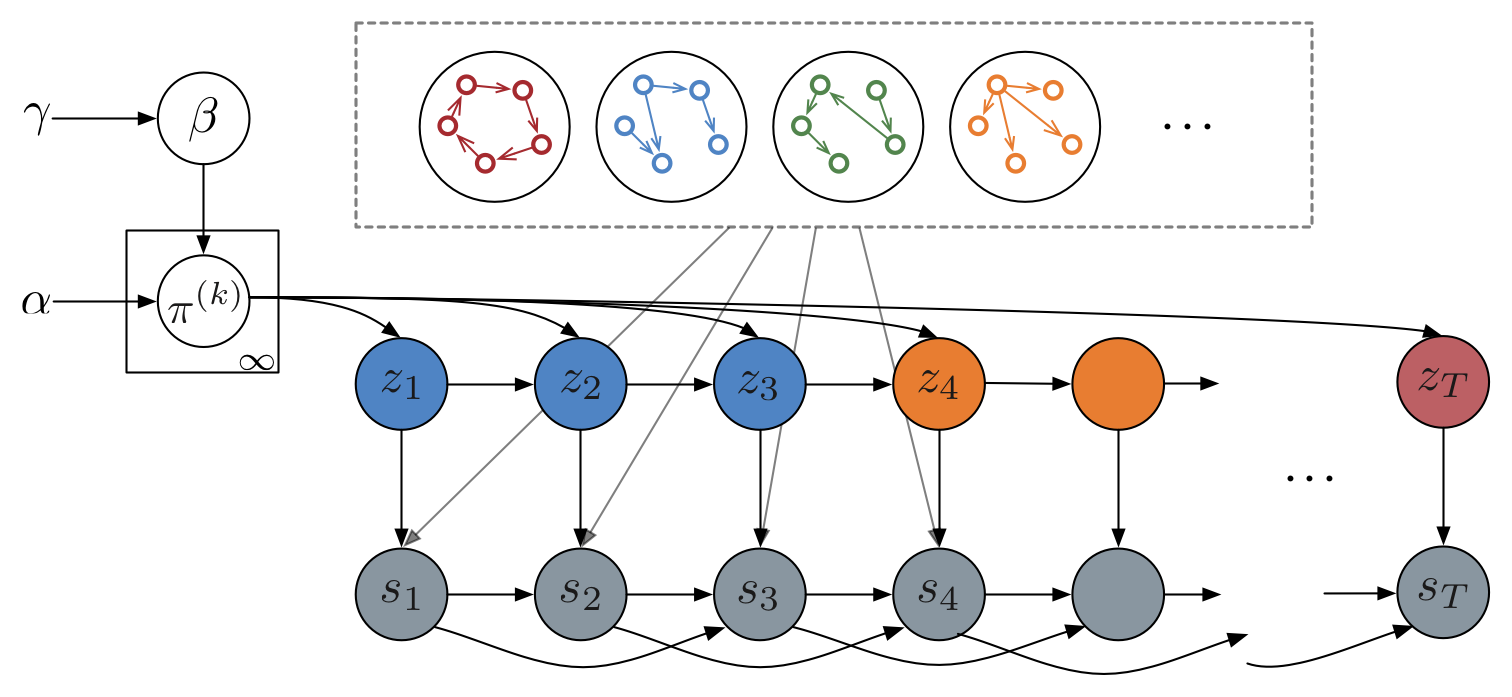
Recurrent switching linear dynamical systems
Many natural systems, such as neurons firing in the brain or basketball teams traversing a court, give rise to time series data with complex, nonlinear dynamics. We can gain insight into these systems by decomposing the data into segments that are each explained by simpler dynamic units. Building on switching linear dynamical systems (SLDS), we present a new model class that not only discovers these dynamical units, but also explains how their switching behavior depends on observations or continuous latent states. These "recurrent" switching linear dynamical systems provide further insight by discovering the conditions under which each unit is deployed, something that traditional SLDS models fail to do. We leverage recent algorithmic advances in approximate inference to make Bayesian inference in these models easy, fast, and scalable. Scott Linderman*, Matthew Johnson*, Andrew C. Miller, Ryan P. Adams, David M. Blei, Liam Paninski,AISTATS 2017 preprint |
|
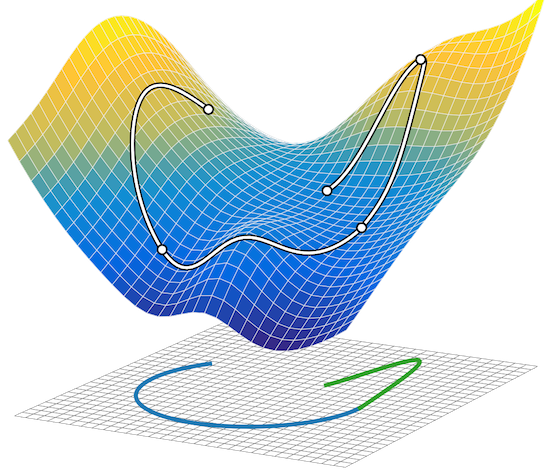
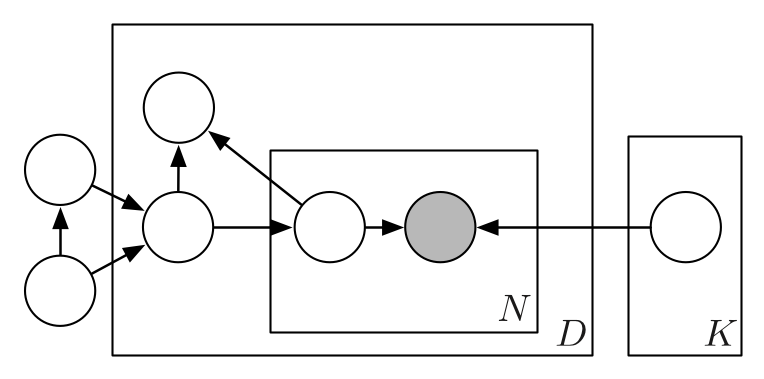
Composing graphical models with neural networks for structured representations and fast inference
We propose a general modeling and inference framework that composes probabilistic graphical models with deep learning methods and combines their respective strengths. Our model family augments graphical structure in latent variables with neural network observation models. For inference, we extend variational autoencoders to use graphical model approximating distributions with recognition networks that output conjugate potentials. All components of these models are learned simultaneously with a single objective, giving a scalable algorithm that leverages stochastic variational inference, natural gradients, graphical model message passing, and the reparameterization trick. We illustrate this framework with several example models and an application to mouse behavioral phenotyping. Matthew Johnson, David Duvenaud, Alex Wiltschko, Bob Datta, Ryan P. AdamsNIPS 2016 arXiv version | NIPS version | NIPS poster | code | bibtex |
|
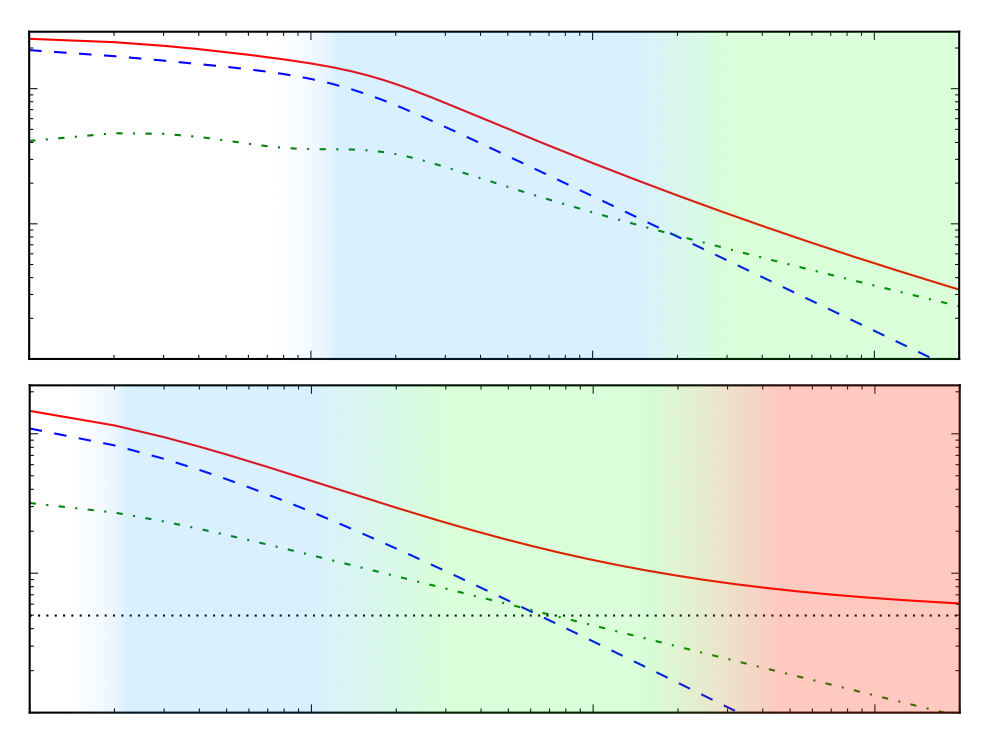
Patterns of scalable Bayesian inference
In this paper, we seek to identify unifying principles, patterns, and intuitions for scaling Bayesian inference. We review existing work on utilizing modern computing resources with both MCMC and variational approximation techniques. From this taxonomy of ideas, we characterize the general principles that have proven successful for designing scalable inference procedures and comment on the path forward. Matthew Johnson*, Elaine Angelino*, Ryan P. Adams(* authors contributed equally; I wrote most of Chapters 2, 5, and 6) Foundations and Trends in Machine Learning Vol. 9, November 2016 preprint (update coming soon) | bibtex |
|
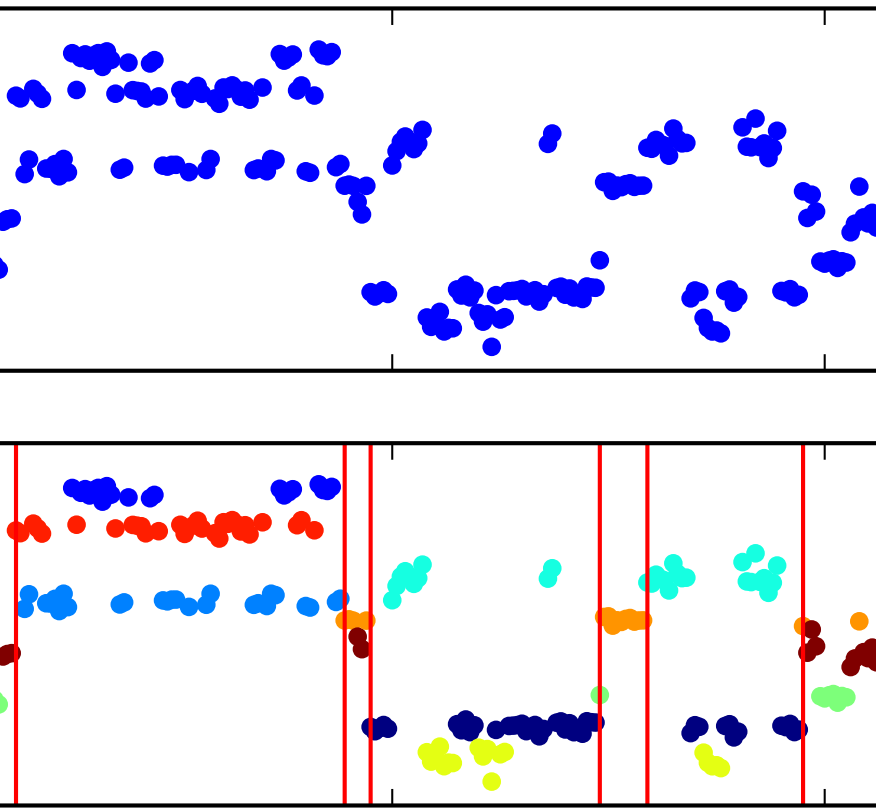
The Segmented iHMM: a simple, efficient hierarchical infinite HMM
We propose the segmented iHMM (siHMM), a hierarchical infinite hidden Markov model (iHMM) that supports a simple, efficient inference scheme. The siHMM is well suited to segmentation problems, where the goal is to identify points at which a time series transitions from one relatively stable regime to a new regime. Conventional iHMMs often struggle with such problems, since they have no mechanism for distinguishing between high- and low-level dynamics. Ardavan Saeedi, Matthew D. Hoffman, Matthew Johnson, Ryan P. AdamsICML 2016 arXiv version | ICML version | bibtex |
|
A Bayesian nonparametric approach for uncovering rat hippocampal population codes during spatial navigation
Rodent hippocampal population codes represent important spatial information about the environment during navigation. We propose an unsupervised Bayesian nonparametric approach to infer these codes from neural spiking and behavior data. To tackle the model selection problem, we apply a hierarchical Dirichlet process-hidden Markov model (HDP-HMM) using two Bayesian inference methods, one based on Markov chain Monte Carlo (MCMC) and the other based on variational Bayes (VB). Scott Linderman, Matthew Johnson, Matthew Willson, Zhe ChenJournal of Neuroscience Methods 2016 paper | bibtex |
|
Cross-corpora unsupervised learning of trajectories in autism spectrum disorders
Patients with developmental disorders, such as autism spectrum disorder (ASD), present with symptoms that change with time even if the named diagnosis remains fixed. We present an unsupervised approach for learning disease trajectories from incomplete medical records combined with disease descriptions from alternate data sources like online forums. In particular, we use a dynamic topic model approach and leverage a recent Polya-gamma augmentation scheme for multinomial observations. We learn disease trajectories from the electronic health records of 13,435 patients with ASD and the forum posts of 13,743 caretakers of children with ASD. Melih Elibol, Vincent Nguyen, Scott Linderman, Matthew Johnson, Amna Hashmi,JMLR 2016 preprint |

|
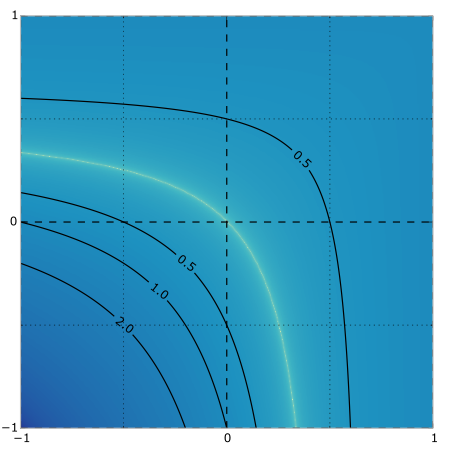
Dependent multinomial models made easy: stick-breaking with the Pólya-gamma augmentation
When modeling categorical and multinomial data it's useful to express dependency structure using latent Gaussian processes, but leaving behind convenient Dirichlet-multinomial conjugacy can result in inefficient or bespoke inference algorithms. Using a logistic stick-breaking representation and the recent Polya-gamma augmentation, we develop an auxiliary variable trick to make inference in these models easy and efficient. In particular, we show how our augmentation allows correlated and dynamic topic models, linear dynamical system models for text, and spatiotemoral count models to leverage off-the-shelf code for GPs and Gaussian linear dynamical systems. Matthew Johnson*, Scott Linderman*, Ryan P. Adams(* authors contributed equally) NIPS 2015 NIPS paper | arXiv version | code | bibtex |
|
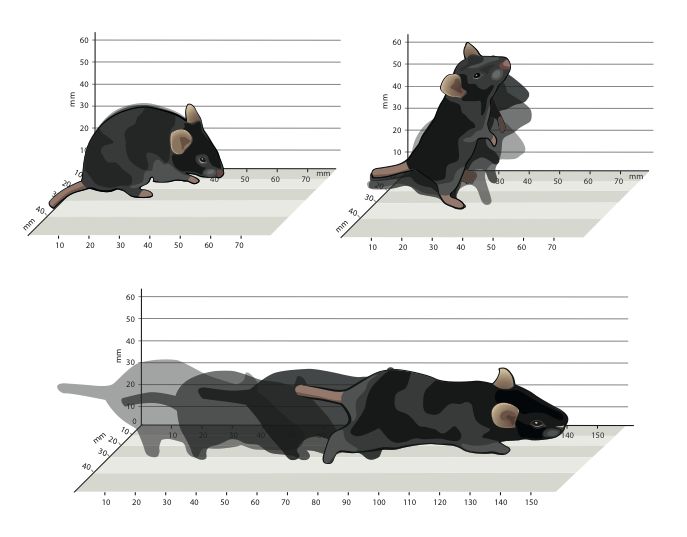
Mapping sub-second structure in mouse behavior
Complex animal behaviors are likely built from simpler modules, but their systematic identification in mammals remains a significant challenge. Here we use depth imaging to show that 3D mouse pose dynamics are structured at the sub-second timescale. Computational modeling of these fast dynamics effectively describes mouse behavior as a series of reused and stereotyped modules with defined transition probabilities. We demonstrate this combined 3D imaging and machine learning method can be used to unmask potential strategies employed by the brain to adapt to the environment, to capture both predicted and previously hidden phenotypes caused by genetic or neural manipulations, and to systematically expose the global structure of behavior within an experiment. This work reveals that mouse body language is built from identifiable components and is organized in a predictable fashion; deciphering this language establishes an framework for characterizing the influence of environmental cues, genes and neural activity on behavior. Alexander B. Wiltschko, Matthew Johnson, Giuliano Iurilli, Ralph E. Peterson, Jesse M. Katon, Stan L. Pashkovski, Victoria E. Abraira, Ryan P. Adams, Sandeep Robert DattaNeuron 2015 paper | video | bibtex |
|
Bayesian time Series models and scalable inference
The main body chapters are broken out below. The background has a concise overview of graphical models. Chapters 4 and 7 have new results that haven't appeared in papers yet.
MIT PhD Thesis, May 2014 thesis | bibtex |
|
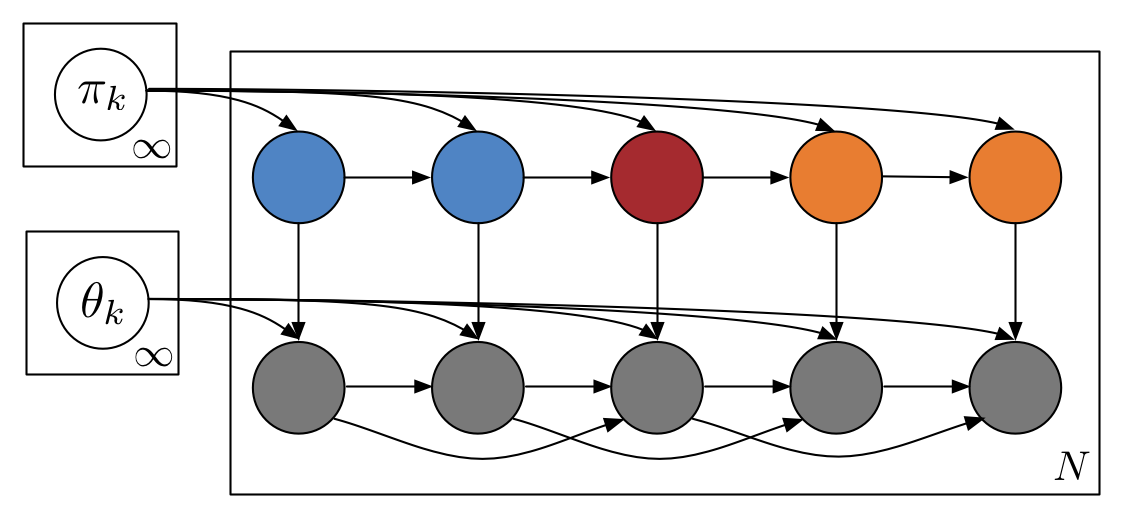
Stochastic variational inferece for Bayesian time series models
Bayesian models provide powerful tools for analyzing complex time series data, but performing inference with large datasets is a challenge. In this paper we develop natural gradient stochastic variational inference (SVI) algorithms for several common Bayesian time series models, namely the hidden Markov model (HMM), hidden semi-Markov model (HSMM), and the nonparametric HDP-HMM and HDP-HSMM. In addition, because HSMM inference can be expensive even in the minibatch setting of SVI, we develop fast approximate updates for some HSMM duration models. Matthew Johnson, Alan S. WillskyICML 2014 paper | code | bibtex |
|
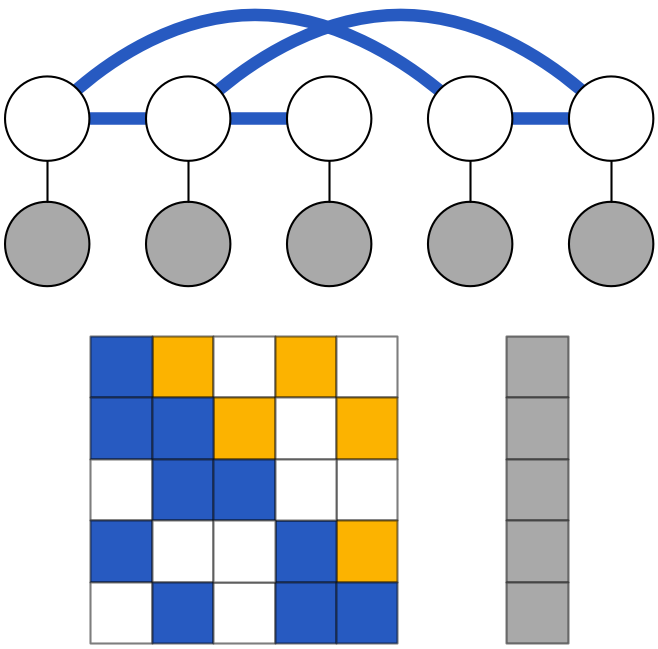
Analyzing Hogwild parallel Gaussian Gibbs sampling
Sampling inference methods are computationally difficult to scale for many models in part because global dependencies can reduce opportunities for parallel computation. Without strict conditional independence structure among variables, standard Gibbs sampling theory requires sample updates to be performed sequentially, even if dependence between most variables is not strong. Empirical work has shown that some models can be sampled effectively by going “Hogwild” and simply running Gibbs updates in parallel with only periodic global communication, but the successes and limitations of such a strategy are not well understood. As a step towards such an understanding, we study the Hogwild Gibbs sampling strategy in the context of Gaussian graphical models. Matthew Johnson, James Saunderson, Alan S. WillskyNIPS 2013 paper | expanded thesis chapter | supplemental | code | bibtex |
|
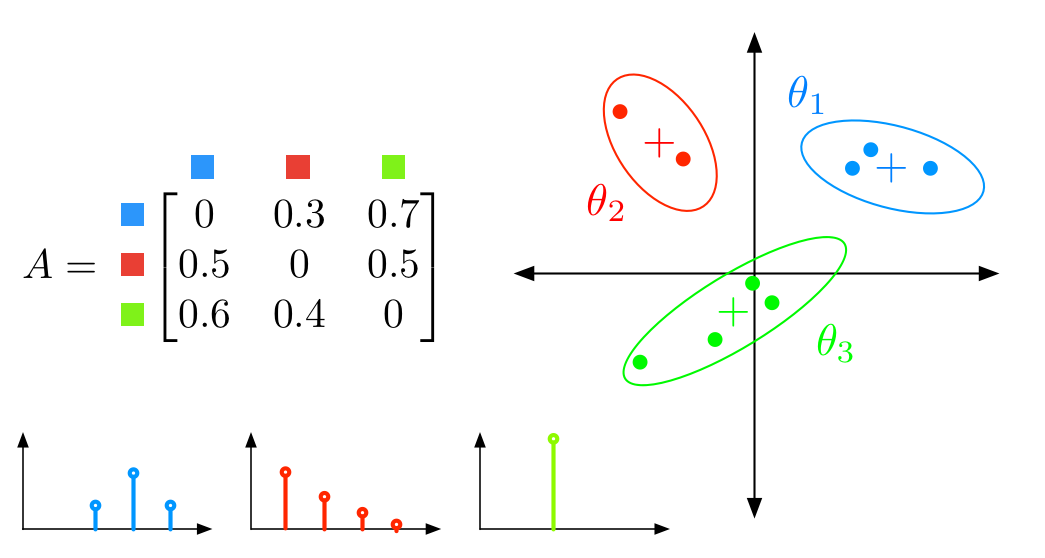
Bayesian nonparametric hidden semi-Markov models
The Hierarchical Dirichlet Process Hidden Markov Model (HDP-HMM) is a natural Bayesian nonparametric extension of the ubiquitous Hidden Markov Model for learning from sequential and time-series data. However, in many settings the HDP-HMM’s strict Markovian constraints are undesirable, particularly if we wish to learn or encode non-geometric state durations. We can extend the HDP-HMM to capture such structure by drawing upon explicit-duration semi-Markov modeling to allow construction of highly interpretable models that admit natural prior information on state durations. In this paper we introduce the explicit-duration Hierarchical Dirichlet Process Hidden semi-Markov Model (HDP-HSMM) and develop sampling algorithms for efficient posterior inference. Matthew Johnson, Alan S. WillskyJMLR 2013 paper | code | bibtex |
|
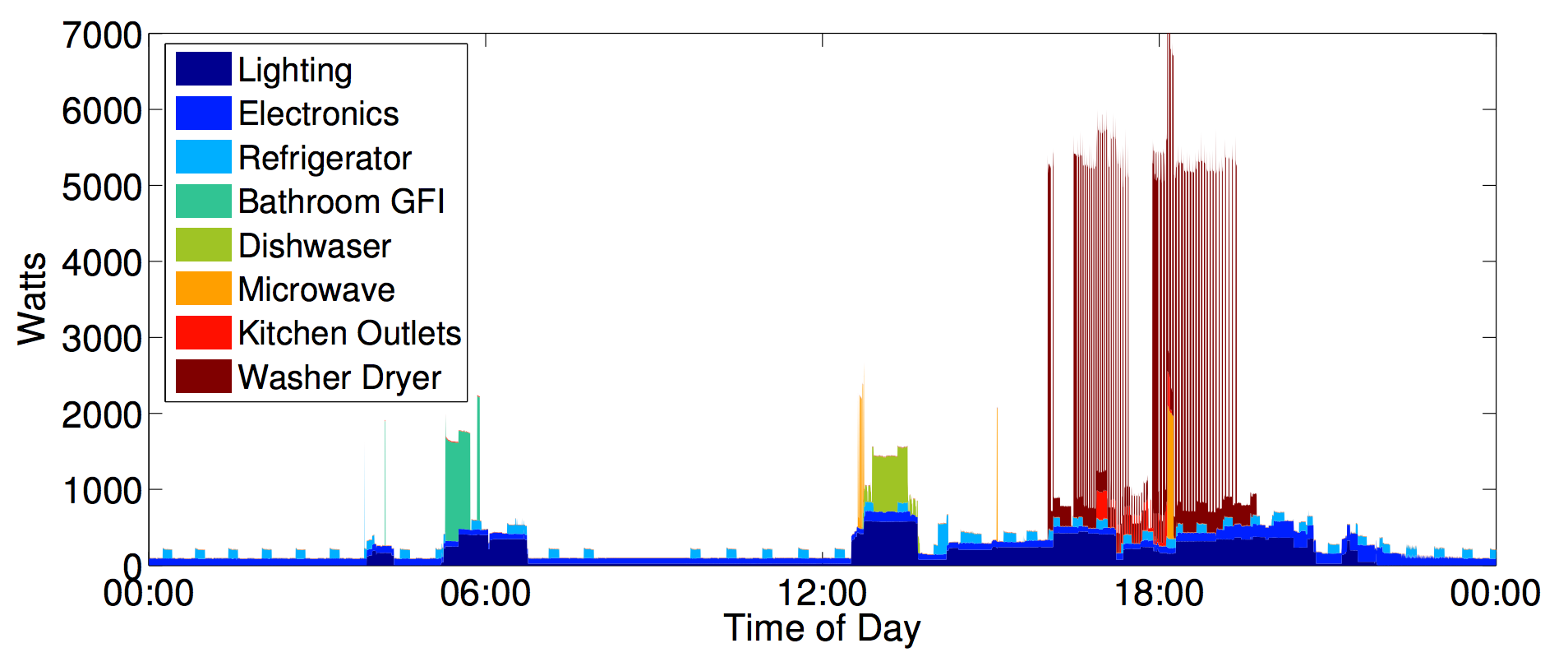
REDD: a public data set for energy disaggregation research
Energy and sustainability issues raise a large number of problems that can be tackled using approaches from data mining and machine learning, but traction of such problems has been slow due to the lack of publicly available data. In this paper we present the Reference Energy Disaggregation Data Set (REDD), a freely available data set containing detailed power usage information from several homes, which is aimed at furthering research on energy disaggregation (the task of determining the component appliance contributions from an aggregated electricity signal). We discuss past approaches to disaggregation and how they have influenced our design choices in collecting data, we describe the hardware and software setups for the data collection, and we present initial benchmark disaggregation results using a well-known Factorial Hidden Markov Model (FHMM) technique. J. Zico Kolter, Matthew JohnsonSustKDD Workshop on Data Mining and Applications in Sustainability 2011 paper | website | bibtex |
|
The hierarchical Dirichlet process hidden semi-Markov model
This was an earlier conference version of the HDP-HSMM paper. Matthew Johnson, Alan S. WillskyUAI 2010 paper | matlab code | bibtex |
|
Necessary and sufficient conditions for high-dimensional salient subset recovery
We consider recovering the salient feature subset for distinguishing between two probability models from i.i.d. samples. Identifying the salient set improves discrimination performance and reduces complexity. The focus in this work is on the high-dimensional regime where the number of variables d, the number of salient variables k and the number of samples n all grow. The definition of saliency is motivated by error exponents in a binary hypothesis test and is stated in terms of relative entropies. It is shown that if n grows faster than max{ck log((d−k)/k), exp(c ′ k)} for constants c, c′ , then the error probability in selecting the salient set can be made arbitrarily small. Thus, n can be much smaller than d. The exponential rate of decay and converse theorems are also provided. An efficient and consistent algorithm is proposed when the dist Vincent Y. F. Tan, Matthew Johnson, Alan S. WillskyISIT 2010 paper | proofs | bibtex |
Abstracts, book chapters, and notes
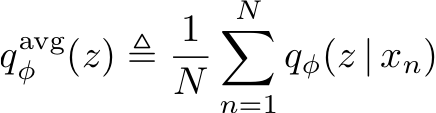
|
ELBO surgery: yet another way to carve up the variational evidence lower bound
We rewrite the variational evidence lower bound objective (ELBO) of variational autoencoders in a way that highlights the role of the encoded data distribution. This perspective suggests that to improve our variational bounds we should improve our priors and not just the encoder and decoder. Matthew D. Hoffman, Matthew JohnsonNIPS 2016 Workshop on Advances in Approximate Bayesian Inference |
|
Bayesian latent state space models of neural activity
Latent state space models such as linear dynamical systems and hidden Markov models are extraordinarily powerful tools for gaining insight into the latent structure underlying neural activity. By beginning with simple hypotheses about the latent states of neural populations and incorporating additional beliefs about the nature of this state and its dynamics, we can compose a nested sequence of increasingly sophisticated models and evaluate them in a statistically rigorous manner. Unfortunately, inferring the latent states and parameters of these models is particularly challenging when presented with discrete spike counts, since the observations are not conjugate with latent Gaussian structure. Thus, we often resort to model-specific approximate inference algorithms which preclude rapid model iteration and typically provide only point estimates of the model parameters. As a result, it is difficult compare models in a way that is robust to the approximation and the particular estimates of the model parameters. Here, we develop a unified framework for composing latent state space models and performing efficient Bayesian inference by leveraging a data augmentation strategy to handle the discrete spike count observations. This framework is easily extensible, as we demonstrate by developing an array of latent state space models with a variety of discrete spike count distributions and fitting them to a simultaneously recorded population of hippocampal place cells. Our Bayesian approach yields a posterior distribution over latent states and parameters, which enables robust prediction and principled model comparison. Moreover, we show that our method is at least as fast as alternative approaches in real-world settings. Scott Linderman, Aaron Tucker, Matthew JohnsonComputational and Systems Neuroscience (Cosyne) 2016 abstract |
|
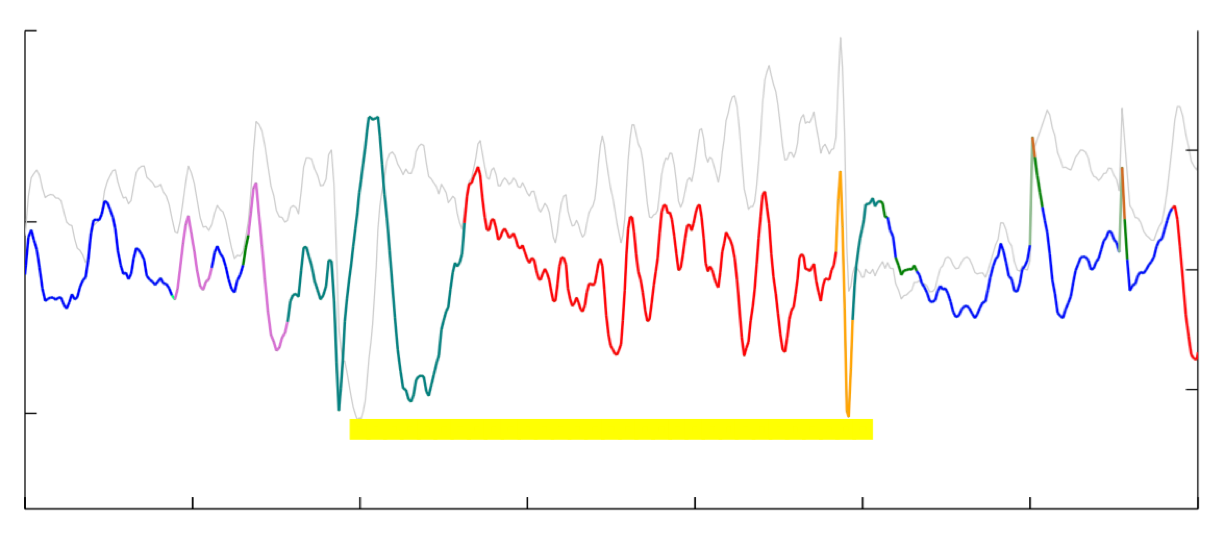
Discovering switching autoregressive dynamics in neural spike train recordings
Generalized linear models (GLM) are powerful tools for identifying dependence in spiking populations of neurons, both over time and within the population Paninski (2004). The GLM identifies these dependencies by modeling spiking patterns through a linear regression and an appropriately-selected link function and likelihood. This regression setup is appealing for its simplicity, the wide variety of available priors, the potential for interpretability, and its computational efficiency. However, the GLM suffers from at least three notable deficiencies. First, the model is linear up to the link function, which only allows a limited range of response maps from neural spiking histories. Second, the model’s parameters are fixed over time, while neural responses may vary due to processes that are exogenous to the population. Third, the generalized linear model presupposes a characteristic time scale for all dynamics, when there may be multiple, varying time scales of neural activity in a given population. Here we seek to address these deficiencies via a switching variant of the generalized linear model. A switching system is one that evolves through a set of discrete states over time, with each state exhibiting its own lowlevel dynamics. For example, the latent state of a hidden Markov model (HMM) can be used to determine the parameters of an autoregressive (AR) process. These HMM-AR models can be used to identify common patterns of linear dependence that vary over time. Bayesian nonparametric versions of HMM-AR models extend these ideas to allow for an infinite number of such patterns to exist a priori, and semi-Markov variants allow the different states to have idiosyncratic duration distributions. Here we develop GLM variants of these switching AR processes and specialize them for neural spiking data. In particular, we exploit recent data augmentation schemes for negative binomial likelihood functions Pillow and Scott (2012) to make inference tractable in HDP-HSMM-AR models with count-based observations. Matthew Johnson Scott Linderman, Sandeep Robert Datta, Ryan P. AdamsComputational and Systems Neuroscience (Cosyne) 2015 abstract |
|
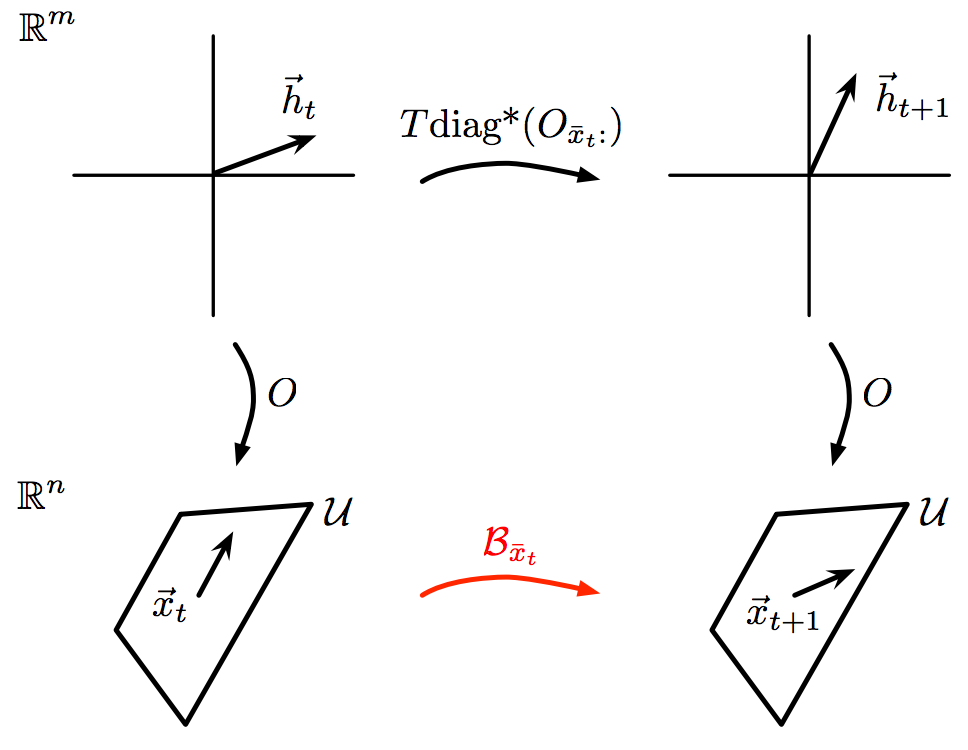
Bayesian nonparametric learning of switching dynamics in cohort physiological time series: application in critical care patient monitoring
The time series of vital signs, such as heart rate (HR) and blood pressure (BP), can exhibit complex dynamic behaviors as a result of internally and externally-induced changes in the state of the underlying control systems. For instance, time series of BP can exhibit oscillations on the order of seconds (e.g., due to the variations in sympathovagal balance), to minutes (e.g., as a conse- quence of fever, blood loss, or behavioral factors), to hours (e.g., due to humoral variations, sleep-wake cycle, or circadian effects) (?, ?). A question of interest is whether “similar” dynamical patterns can be automatically identified across a heterogeneous patient cohort, and be used for prognosis of patients’ health and progress. In this work, we present a Bayesian nonparametric switching Markov processes framework with conditionally linear dynamics to learn phenotypic dynamic behaviors from vital sign time series of a patient cohort, and use the learned dynamics to characterize the changing physiological states of patients for critical-care bed-side monitoring. Li-Wei Lehman, Matthew Johnson, Shamim Nemati, Ryan P. Adams, Roger G. MarkChapter 11 in Advanced State Space Methods for Neural and Clinical Data, CUP 2015 chapter | bibtex |
|
A simple explanation of A Spectral Algorithm for Learning Hidden Markov Models
A simple linear algebraic explanation of the algorithm in "A Spectral Algorithm for Learning Hidden Markov Models" (COLT 2009). Most of the content is in Figure 2; the text just makes everything precise in four nearly-trivial claims. Matthew JohnsonarXiv note |
|
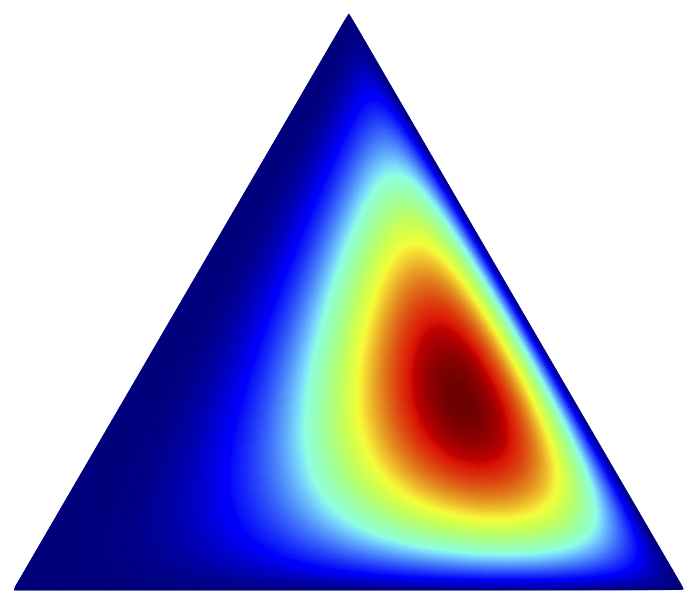
Dirichlet posterior sampling with truncated multinomial likelihoods
We consider the problem of drawing samples from posterior distributions formed under a Dirichlet prior and a truncated multinomial likelihood, by which we mean a Multinomial likelihood function where we condition on one or more counts being zero a priori. Sampling this posterior distribution is of interest in inference algorithms for hierarchical Bayesian models based on the Dirichlet distribution or the Dirichlet process, particularly Gibbs sampling algorithms for the Hierarchical Dirichlet Process Hidden Semi-Markov Model. We provide a data augmentation sampling algorithm that is easy to implement, fast both to mix and to execute, and easily scalable to many dimensions. We demonstrate the algorithm's advantages over a generic Metropolis-Hastings sampling algorithm in several numerical experiments. Matthew Johnson, Alan S. WillskyarXiv note |
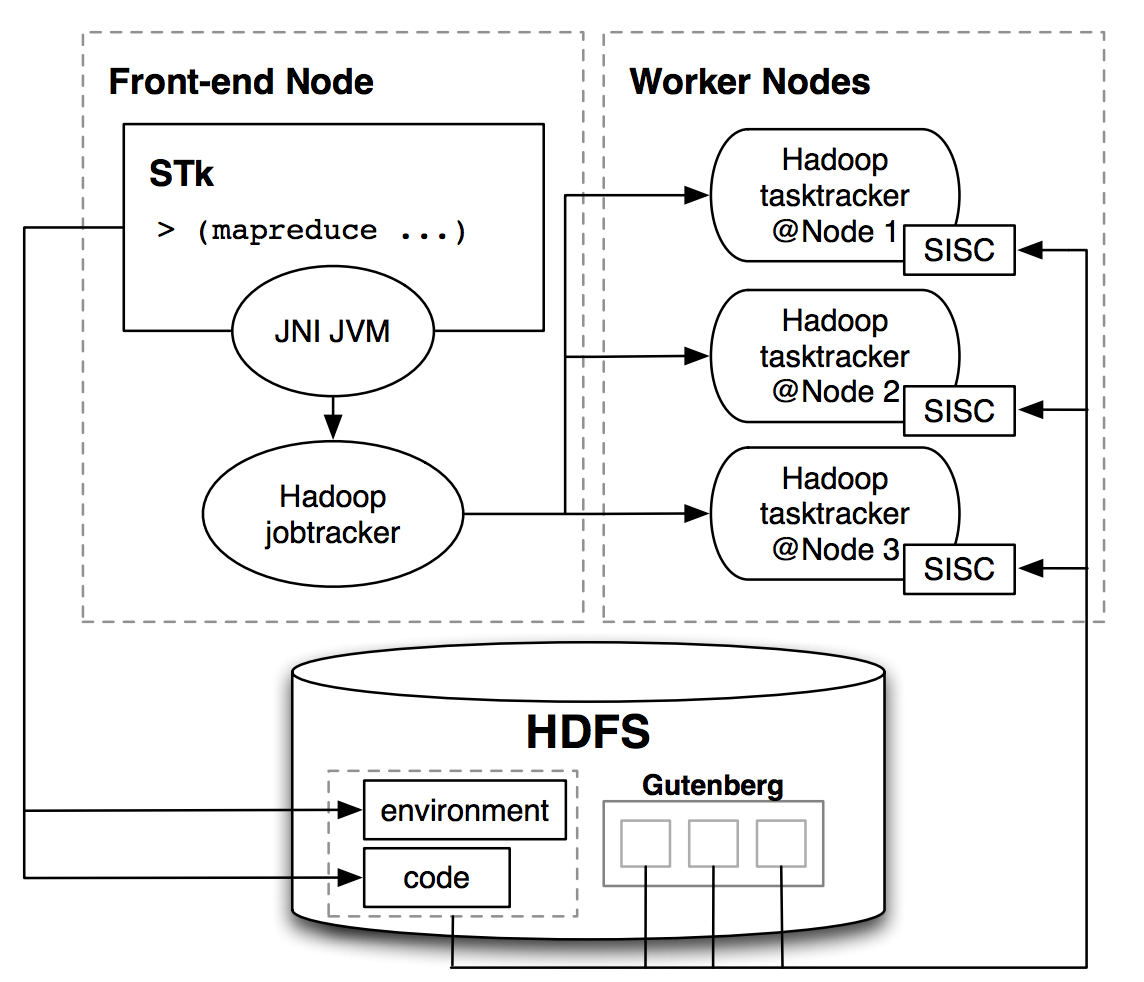
|
Infusing parallelism into introductory computer science curriculum using MapReduce
We have incorporated cluster computing fundamentals into the introductory computer science curriculum at UC Berkeley. For the first course, we have developed coursework and programming problems in Scheme centered around Google’s MapReduce. To allow students only familiar with Scheme to write and run MapReduce programs, we designed a functional interface in Scheme and implemented software to allow tasks to be run in parallel on a cluster. The streamlined interface enables students to focus on programming to the essence of the MapReduce model and avoid the potentially cumbersome details in the MapReduce implementation, and so it delivers a clear pedagogical advantage. The interface’s simplicity and purely functional treatment allows students to tackle data-parallel problems after the first two-thirds of the first introductory course. In this paper we describe the system implementation to interface our Scheme interpreter with a cluster running Hadoop (a Java-based MapReduce implementation). Our design can serve as a prototype for other such interfaces in educational environments that do not use Java and therefore cannot simply use Hadoop. We also outline the MapReduce exercises we have introduced to our introductory course, which allow students in an introductory programming class to begin to work with data-parallel programs and designs. Matthew Johnson*, Ramesh Sridharan*, Robert H. Liao, Alexander Rasmussen, Dan Garcia, Brian K. Harvey(* authors contributed equally) UC Berkeley Tech Report No. UCB/EECS-2008-34, 2008 tech report | bibtex |